Overview
The Gilad lab uses a combination of within-species and between-species perspectives to study gene regulatory evolution in humans and other primates. We leverage the large effect sizes that are typically observed in comparative studies (when one considers differences between species) to identify possible connections between regulatory changes and adaptation. In turn, using the platform of regulatory quantitative trait locus (QTL) mapping and mediation analysis within species, we develop informed hypotheses about the casual relationships that propagate the effects of genetic variation, through changes in molecular mechanisms, to differences in gene regulation and, ultimately to a distinct phenotype.

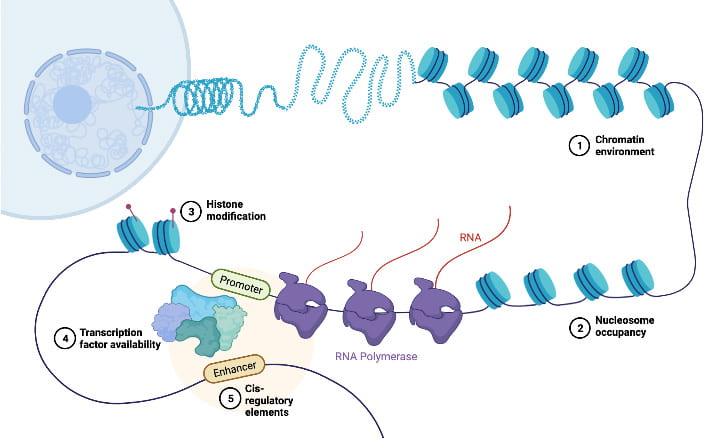
Gene regulatory dynamics and the molecular basis of human traits
Gene regulation is inherently dynamic. A mutation’s effect may emerge only in specific cell types, under particular environmental conditions, or at critical time points during development. We use a variety of strategies and model systmes to capture context-specific genetic effects on gene regulation, which helps us understand how noncoding genetic variants contribute to phenotypic variation between individuals and species. We have established a comparative iPSC panel that includes multiple human and chimpanzee cell lines. These iPSCs are freely available to the scientific community upon request.
High-resolution molecular QTL maps
We use bulk and single-cell sequencing to measure gene expression, chromatin accessibility, splicing, and other molecular traits across tissues, cell types, and individuals, integrating these data to discern the regulatory cascade that connects genetic variation to higher-level functions.
Benchmarking sequencing methods and tools that facilitate the study of alternative polyadenylation
Gene-by-environment interactions in primates
Using our panel of primate stem cells, we study how humans and other primates differ in their responses to external perturbations and treatments. between species.
Response eQTL mapping
Gene activity can change dramatically in response to an external stimulus, and much of this activity is genetically regulated. We measure cell-type-specific responses to disease-relevant perturbations and treatments to identify candidate genotypes, genes, and tissues that mediate disease risk.
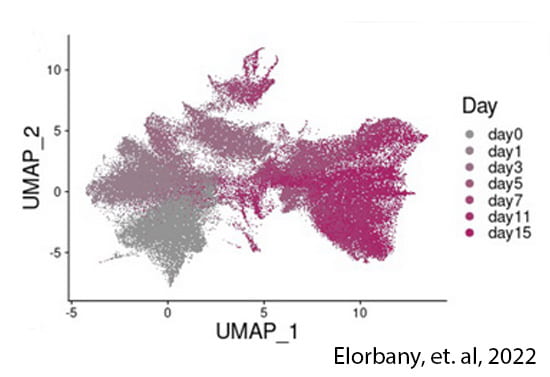
Temporally dynamic gene regulation
Trait-associated variants can have transient effects, emerging only at a critical timepoint or developmental window. We use stem cells to characterize gene regulatory variation during cellular differentiation, both within and between species.
Tools And Models
Enlarge image
Functional Genomics
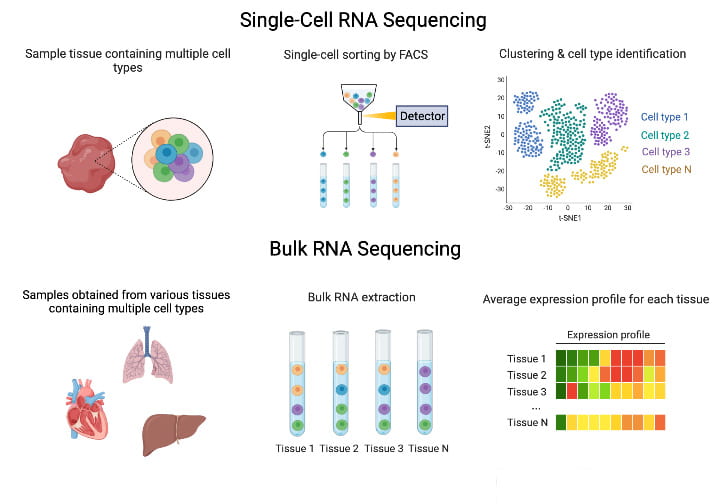
The majority of our experiments use bulk or single-cell RNA sequencing (scRNA-seq). Bulk RNA sequencing is quick and inexpensive, which allows us to survey a variety of tissues, but it lacks the cellular resolution single-cell techniques provide. We also use other molecular sequencing techniques (e.g., ATAC-seq) as needed.
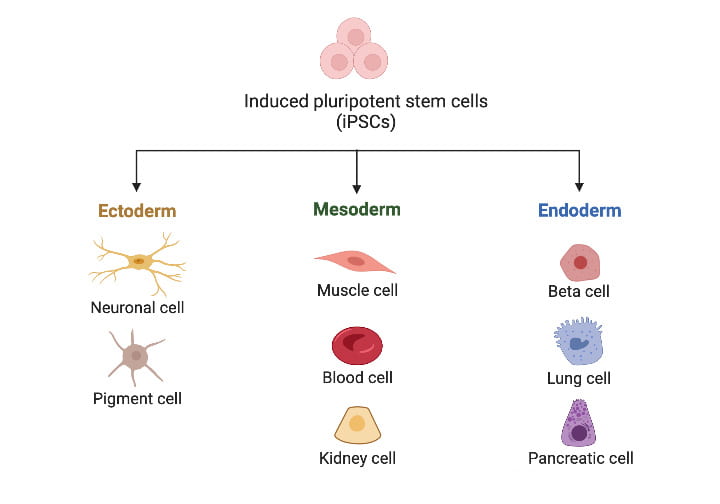
Directed stem cell differentiation
When we intend to study a specific cell type, we use carefully designed protocols to guide the differentiation of our iPSCs. We have successful directed differentiation protocols for many cell types, including cardiomyocytes and chondrocytes.
Enlarge image
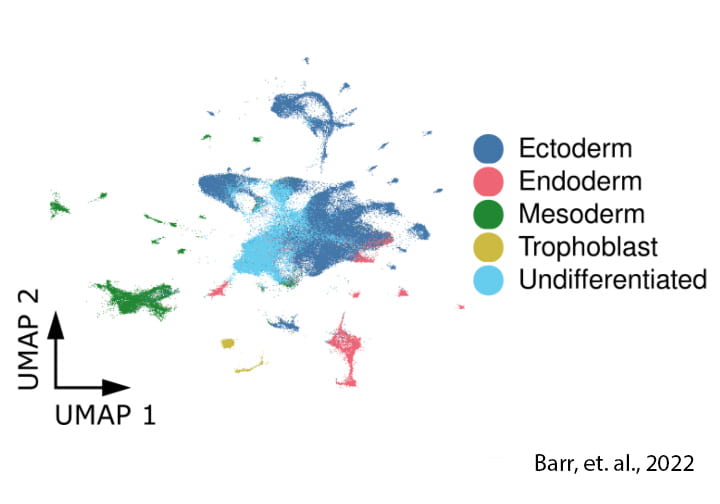
Guided stem cell differentiation
We can also obtain a lot of useful information from iPSCs that are allowed to differentiate spontaneously (guided differentiation). These resulting masses have a high level of cellular heterogeneity and allow us to monitor dynamic regulation in multiple cell types simultaneously.
Unless otherwise indicated, images generated using Biorender.com