Publications
Preprints
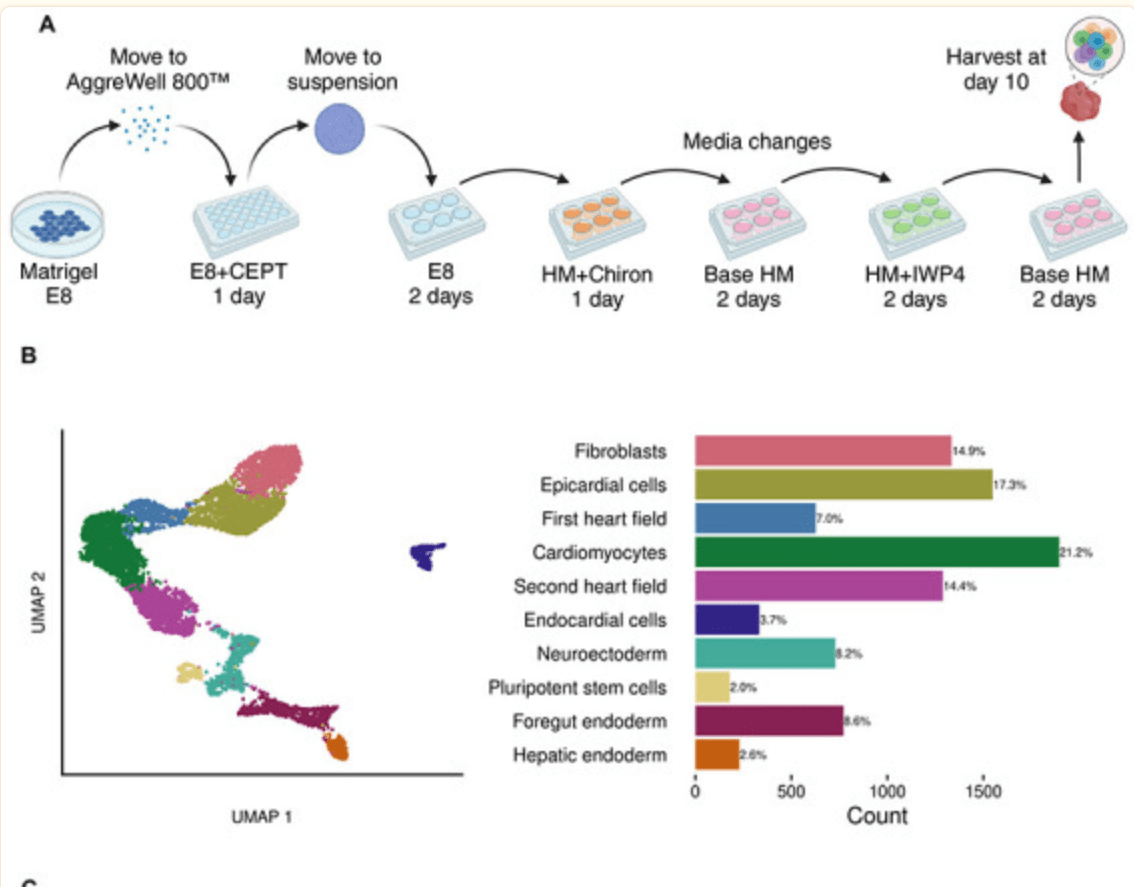
Guided Differentiation of Pluripotent Stem Cells for Cardiac Cell Diversity
McIntire E, Barr KA, Gonzales NM, Gilad Y
bioRxiv; 2024

Widespread conservation of chromatin accessibility patterns and transcription factor binding in human and chimpanzee induced pluripotent stem cells
Gallego Romero I, Gopalakrishnan, S and Gilad Y
bioRxiv; 2018
2025

Disease-associated loci share properties with response eQTLs under common environmental exposures
Lin W, Li M, Allen O, Burnett J, Popp J, Stevens M, Battle A, Gilad Y.
ResearchSquare; 2025

Oxygen-induced stress reveals context-specific gene regulatory effects in human brain organoids
Umans BD, Gilad Y
Genome Res; 2025

An Intuitive Primer on Effective Functional Genomic Study Design (textbook)
Gilad Yoav
Available on Amazon or Barnes and Noble
2024

Cell type and dynamic state govern genetic regulation of gene expression in heterogeneous differentiating cultures
Popp JM, Rhodes K, Jangi R, Li M, Barr KA, Tayeb K, Battle A, Gilad Y
Cell Genomics; 2024
Genetic regulatory effects in response to a high cholesterol, high fat diet in baboons
Lin W, Wall JD, Li G, Newman D, Yang Y, Abney M, VandeBerg JL, Olivier M, Gilad Y, Cox LA
Cell Genomics; 2024
2023
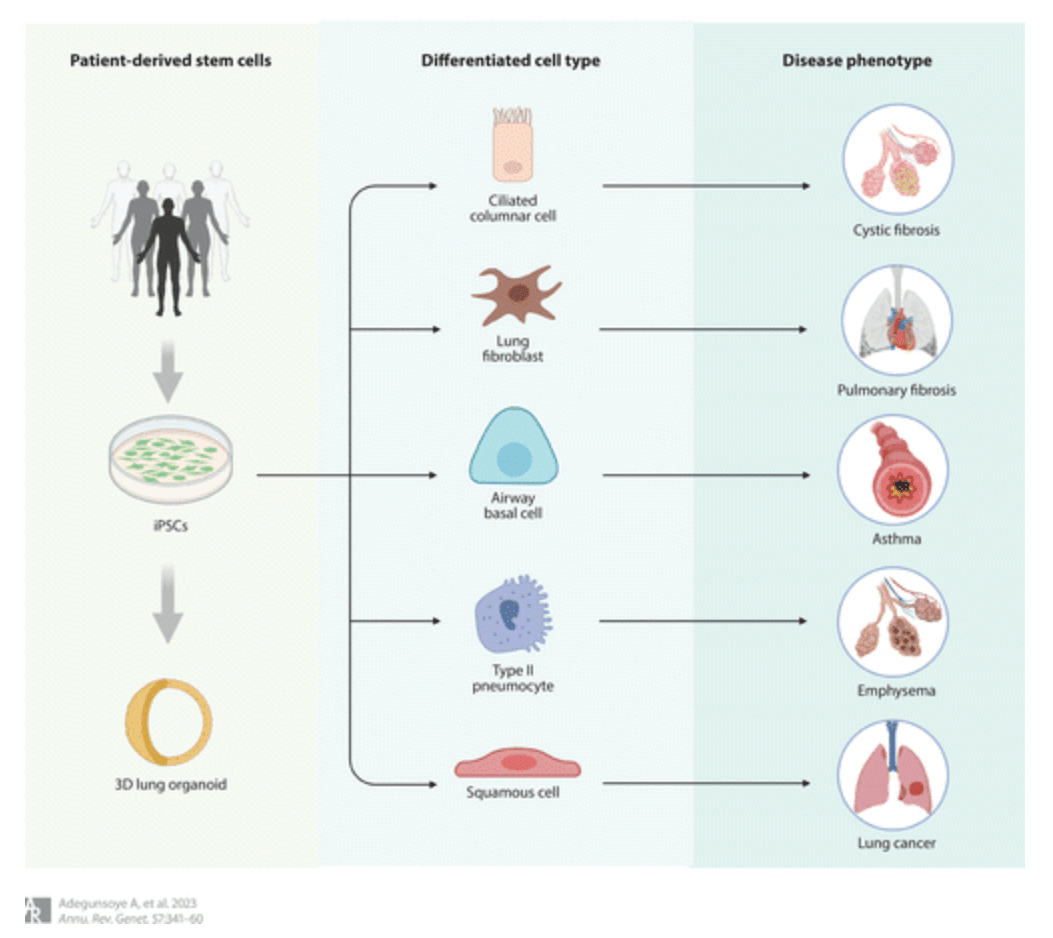
Induced Pluripotent Stem Cells in Disease Biology and the Evidence for Their In Vitro Utility
Adegunsoye, A, Gonzales, NM and Gilad Y
Annu Rev Genet; 2023
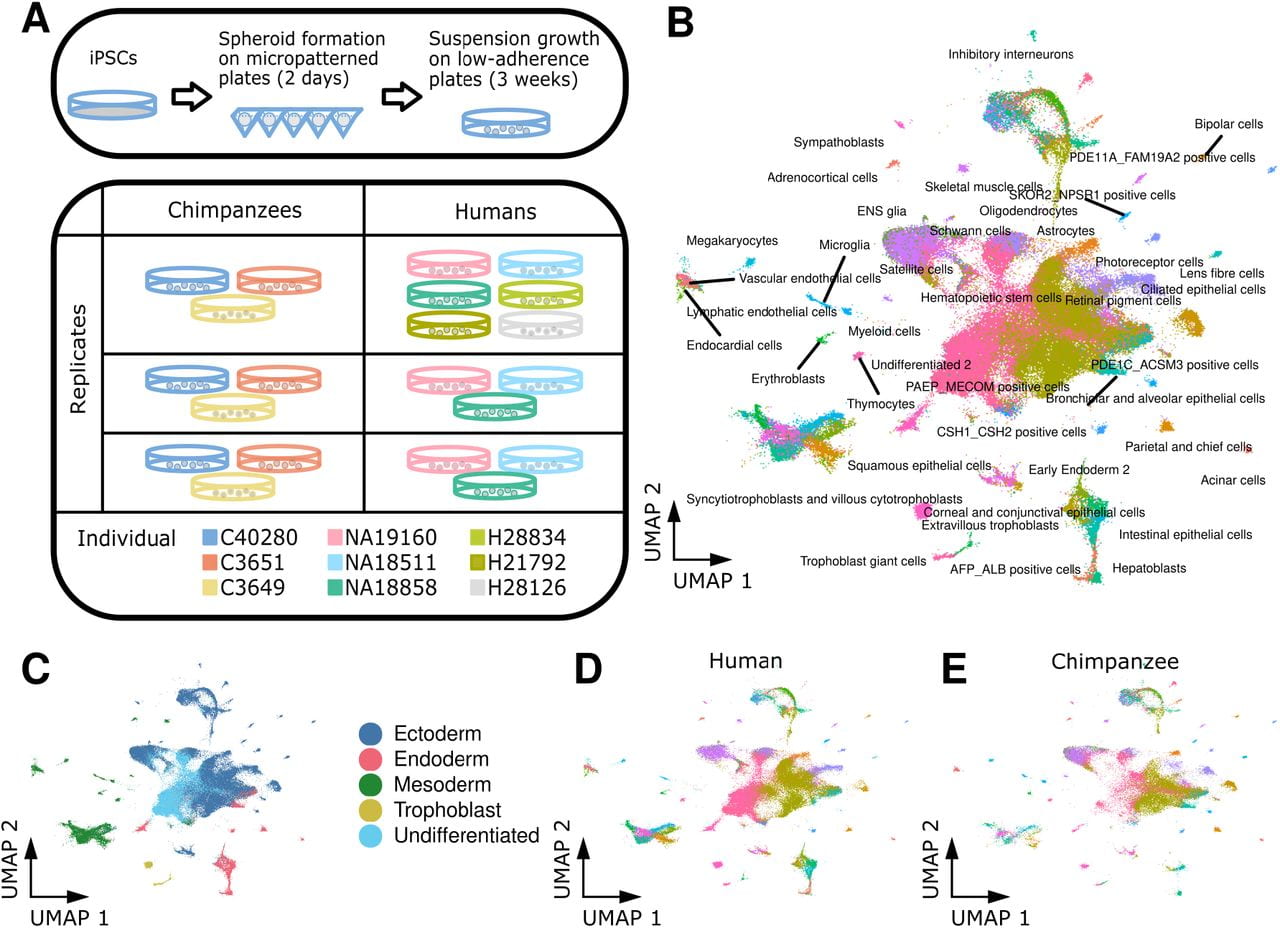
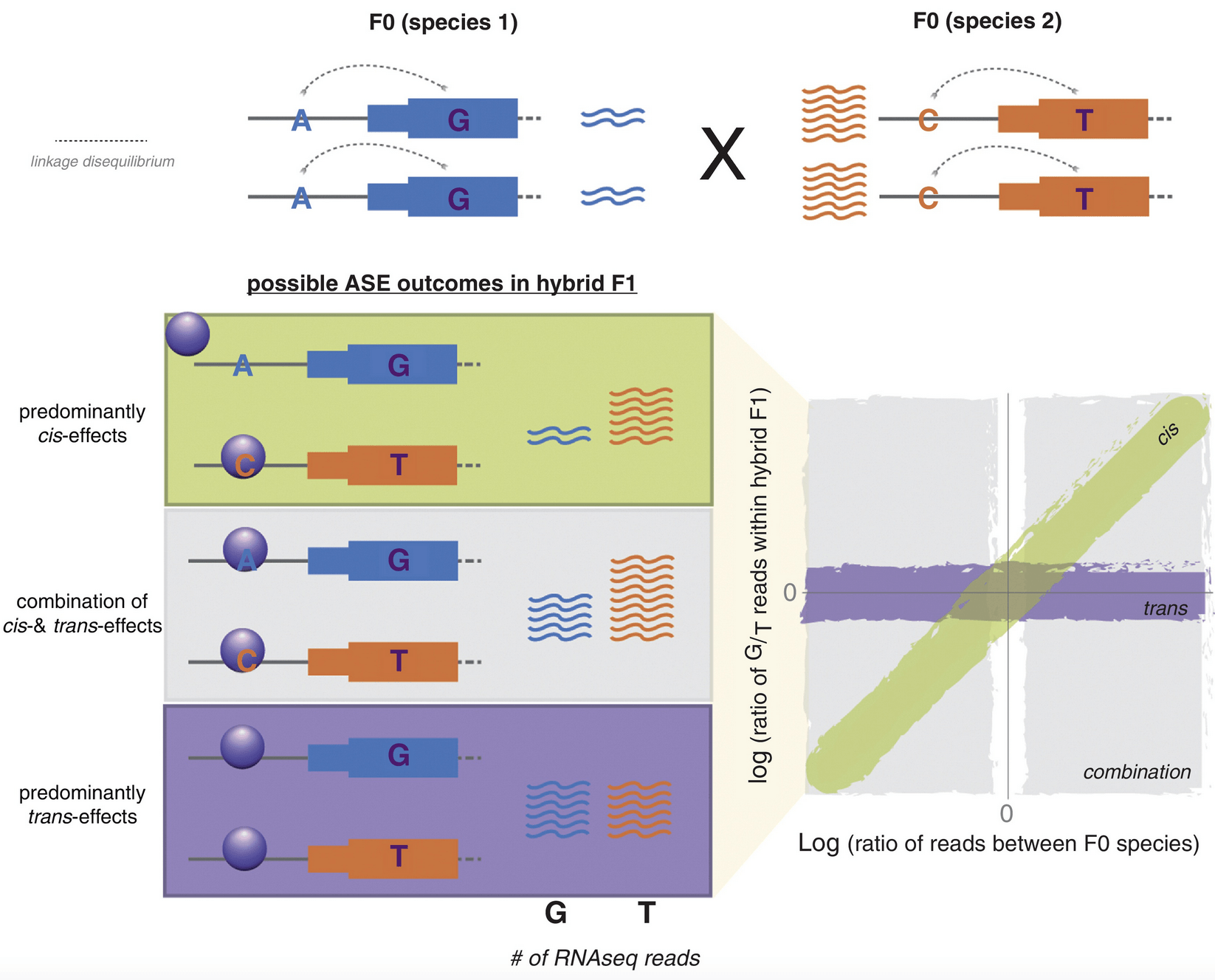
The relationship between regulatory changes in cis and trans and the evolution of gene expression in humans and chimpanzees
Barr KA, Rhodes KL, and Gilad Y
Genome Biology; 2023
2022
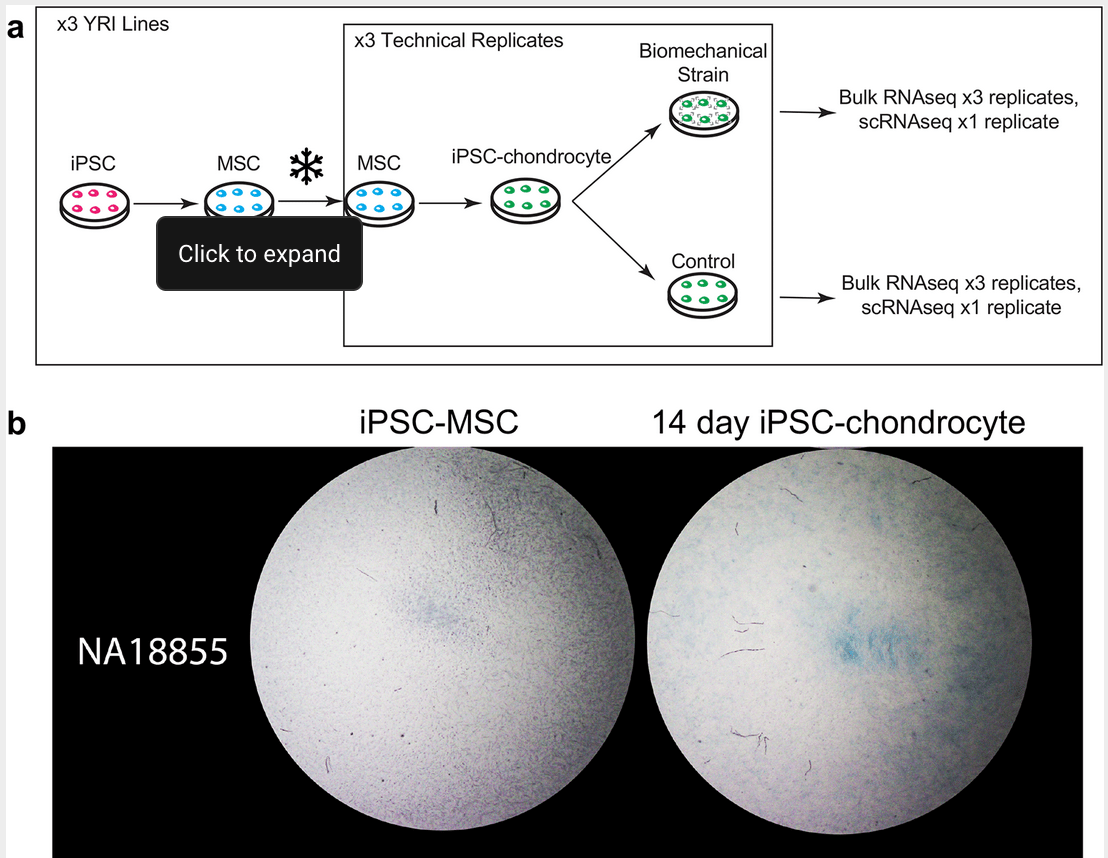
Characterizing gene expression in an in vitro biomechanical strain model of joint health [version 2; peer review: 1 approved, 1 not approved]
Hung A, Housman G, Briscoe EA, Cuevas C and Gilad Y
F1000 Research; 2022
Evolutionary insights into primate skeletal gene regulation using a comparative cell culture model
Housman G, Briscoe E, and Gilad Y
PLoS Genetics; 2022
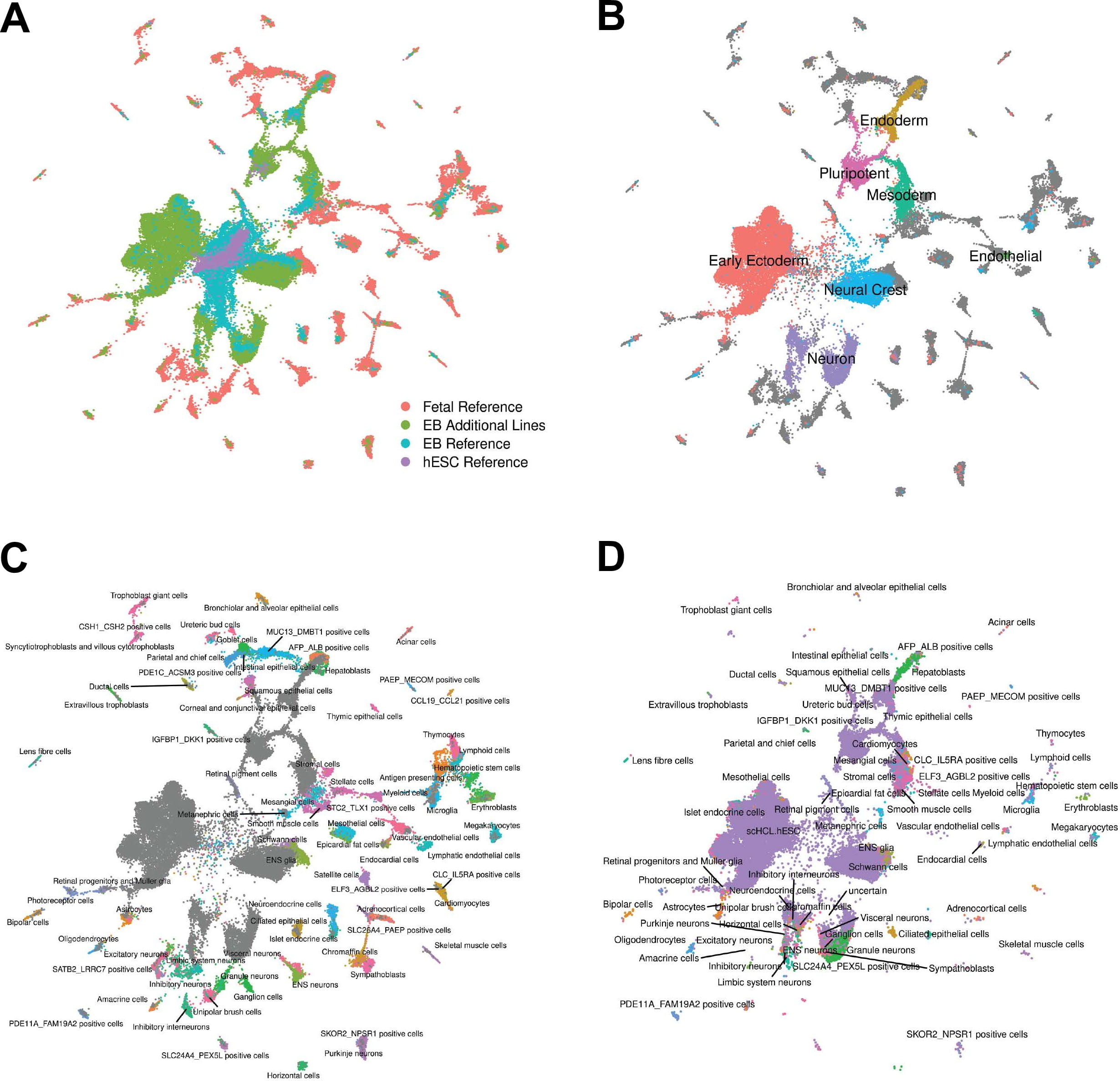
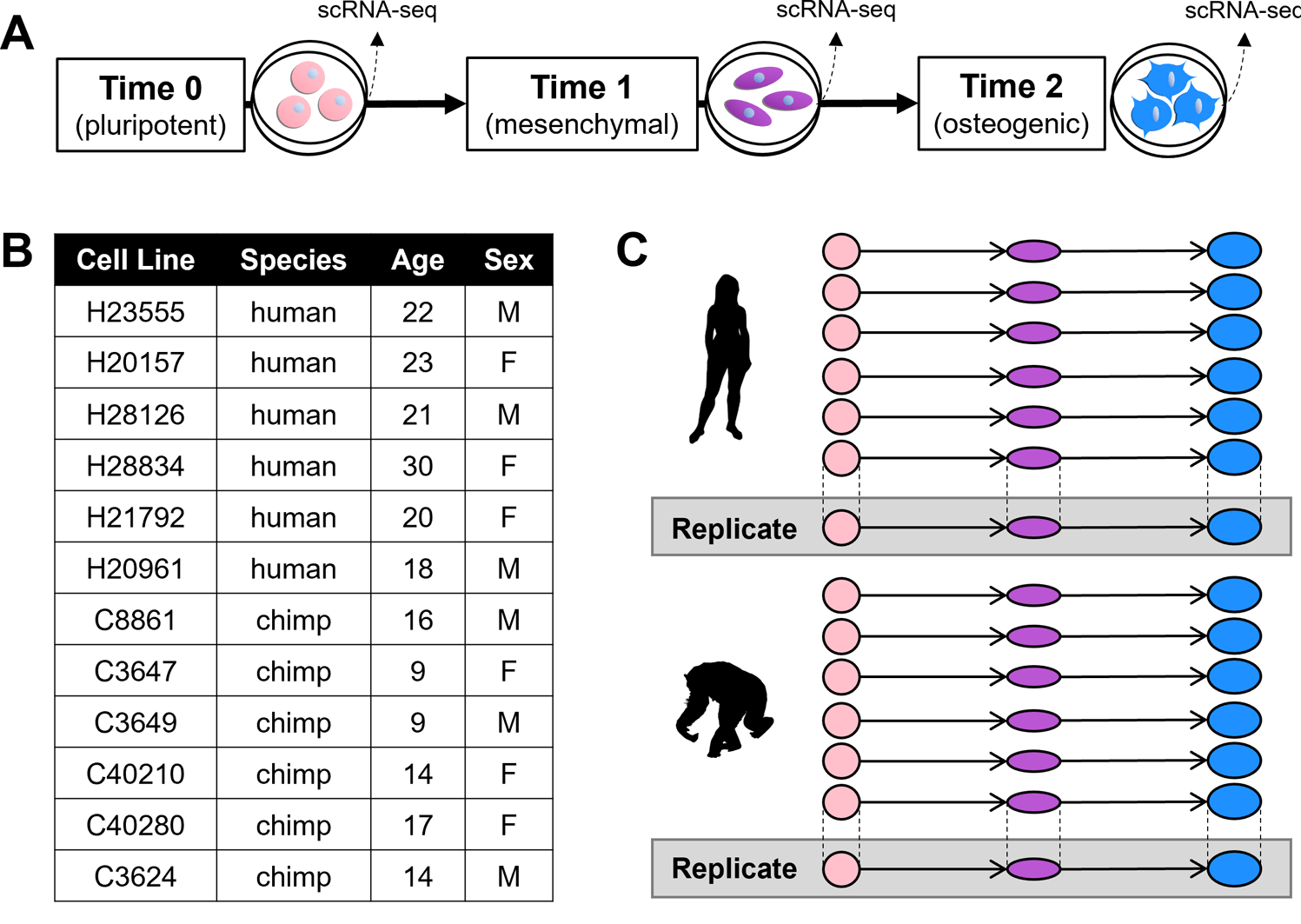
Human embryoid bodies as a novel system for genomic studies of functionally diverse cell types
Rhodes K, Barr KA, Popp JM, Strober BJ, Battle A, and Gilad Y
eLife; 2022
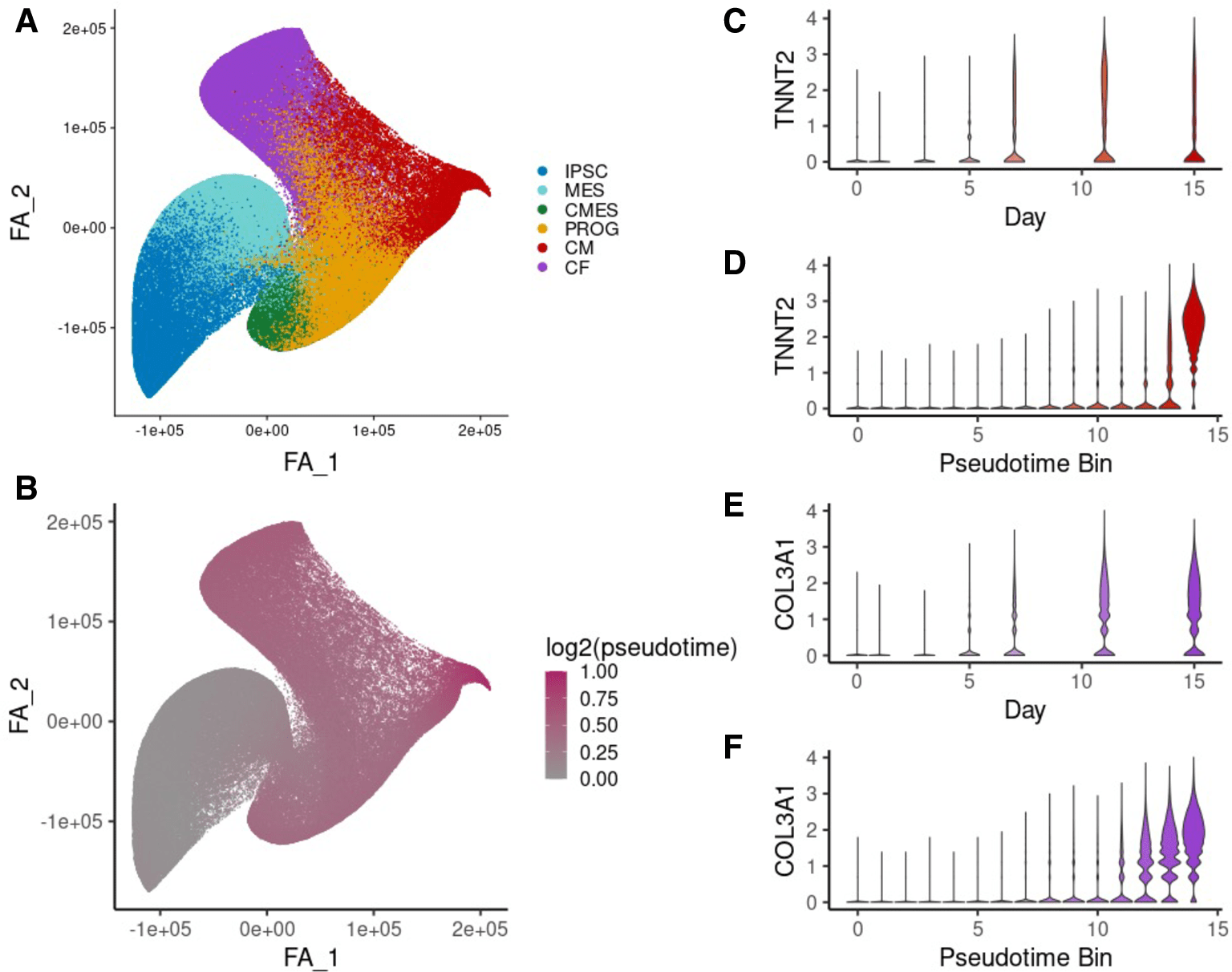
Single-cell sequencing reveals lineage-specific dynamic genetic regulation of gene expression during human cardiomyocyte differentiation
Elorbany R, Popp JM, Rhodes K, Strober BJ, Barr KA, Qi G, Gilad Y, and Battle A
PLoS Genetics; 2022
Supplementary data: upload pending
2021
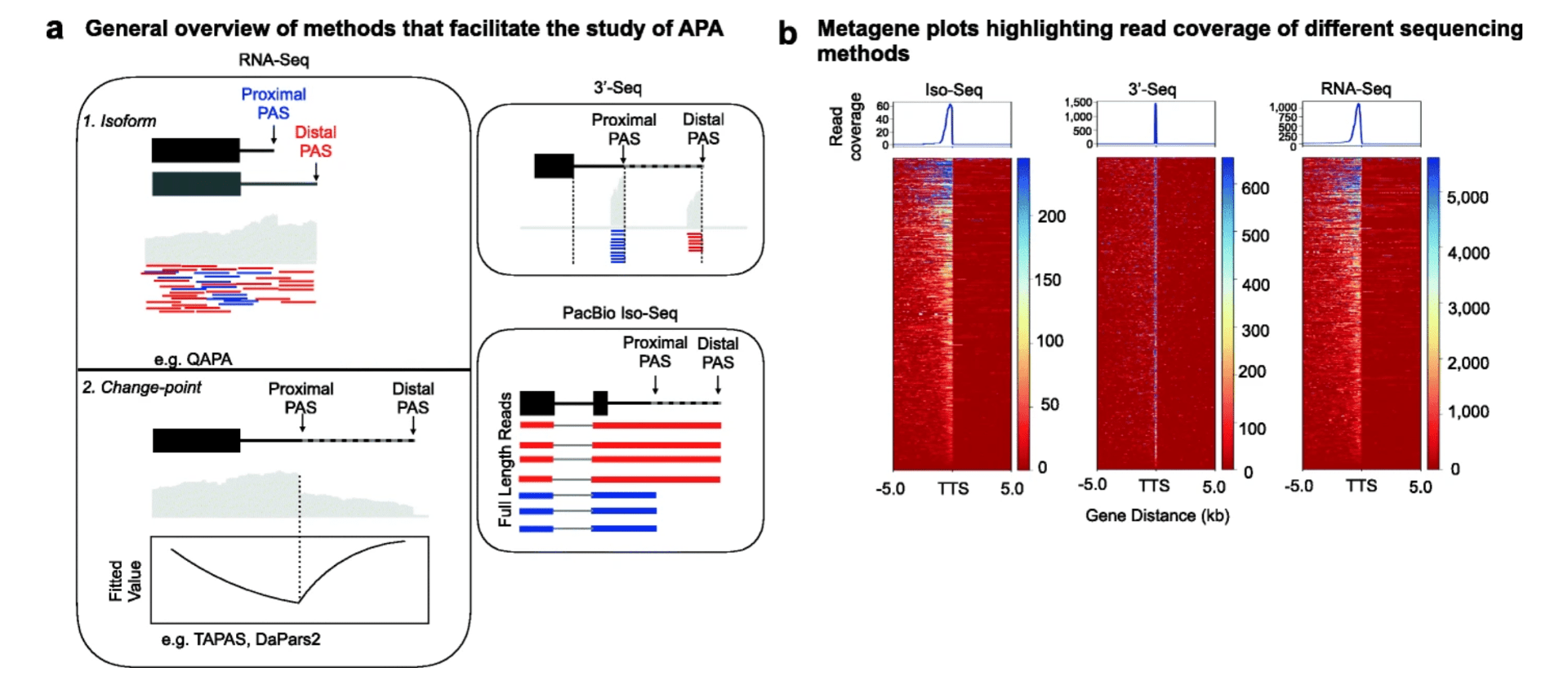
Benchmarking sequencing methods and tools that facilitate the study of alternative polyadenylation
Shah A, Mittleman BE, Gilad Y, and Li YI
Genome Biology; 2021
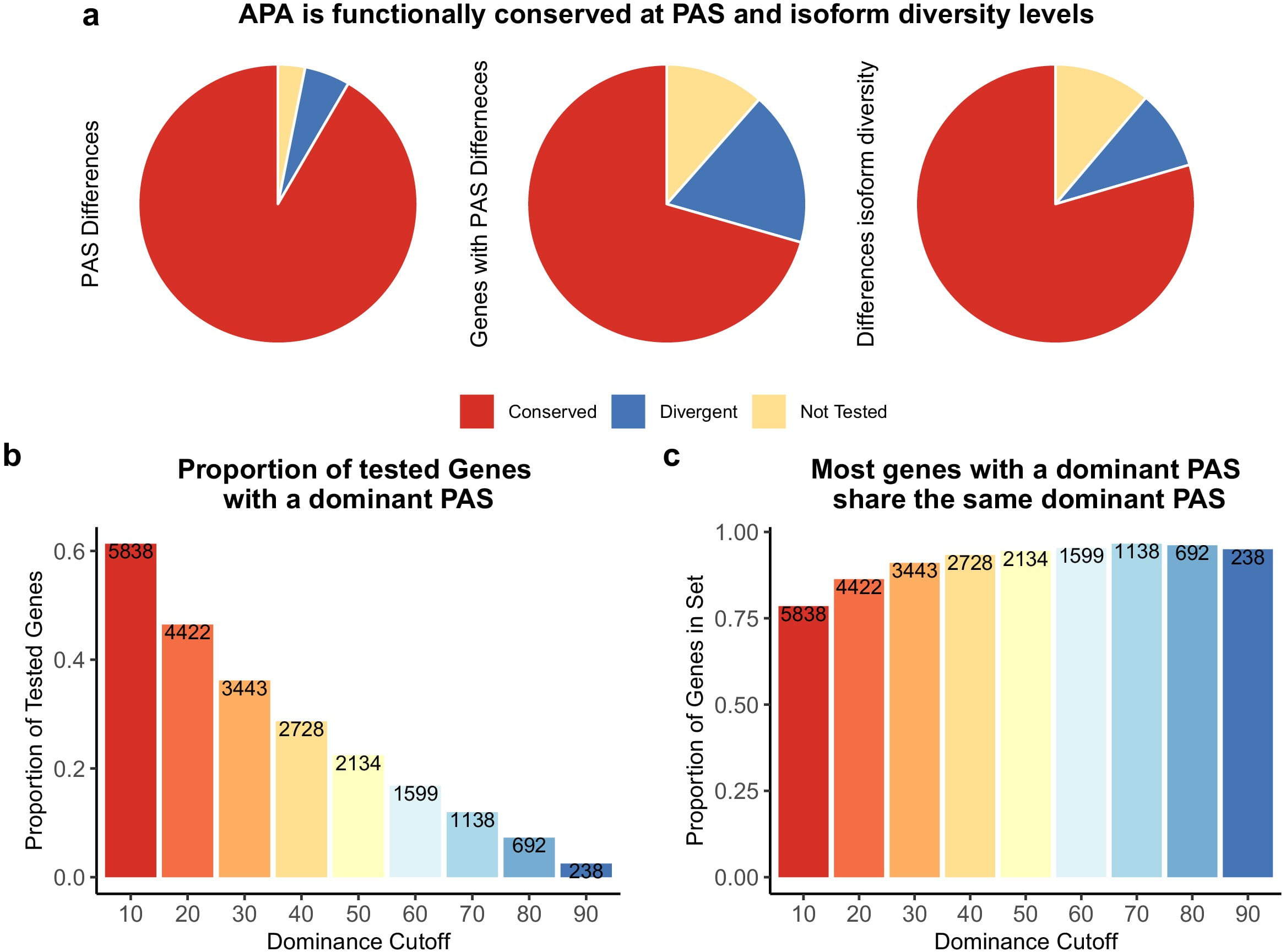
Divergence in alternative polyadenylation contributes to gene regulatory differences between humans and chimpanzees
Mittleman BE, Pott S, Warland S, Barr K, Cuevas C, and Gilad Y
eLife; 2021
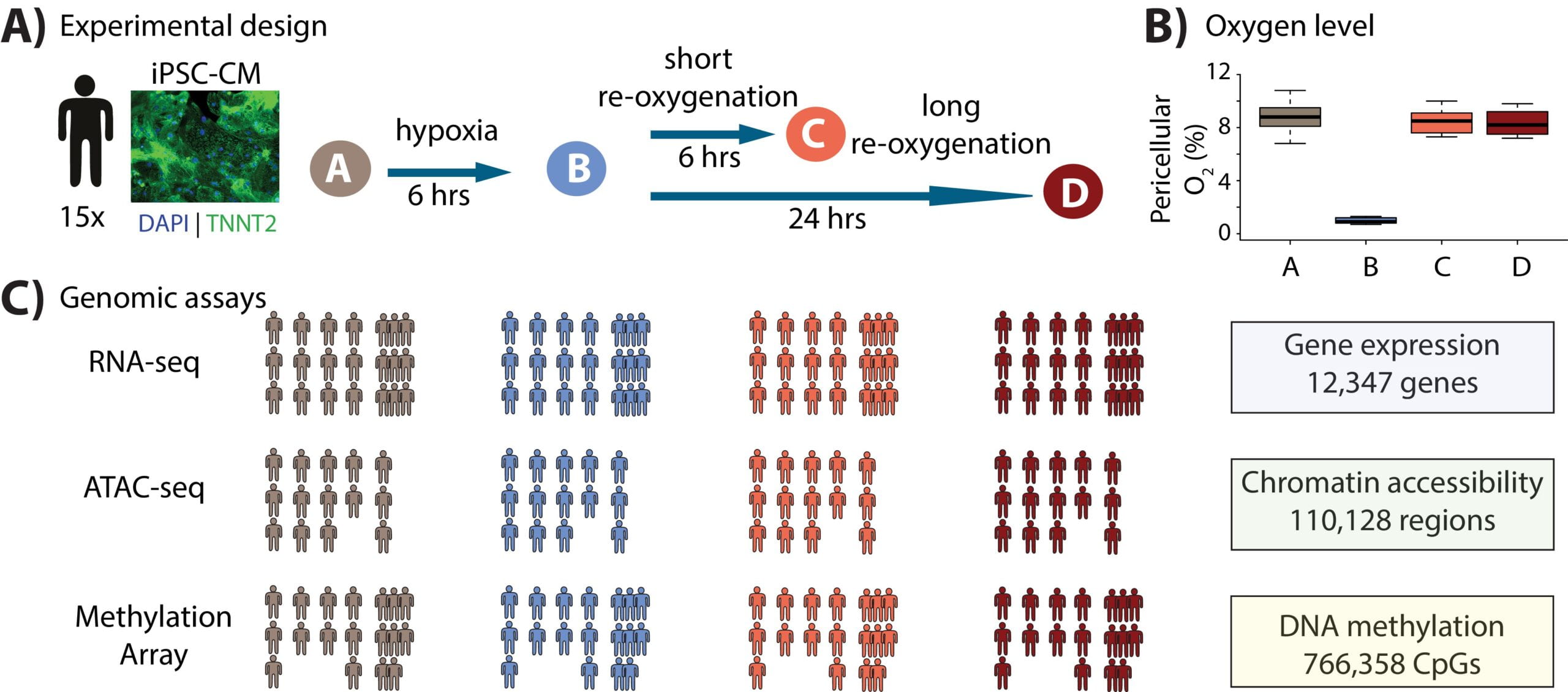
Dynamic effects of genetic variation on gene expression revealed following hypoxic stress in cardiomyocytes
Ward MC, Banovich NE, Sarkar A, Stephens M, and Gilad Y
eLife; 2021

2020
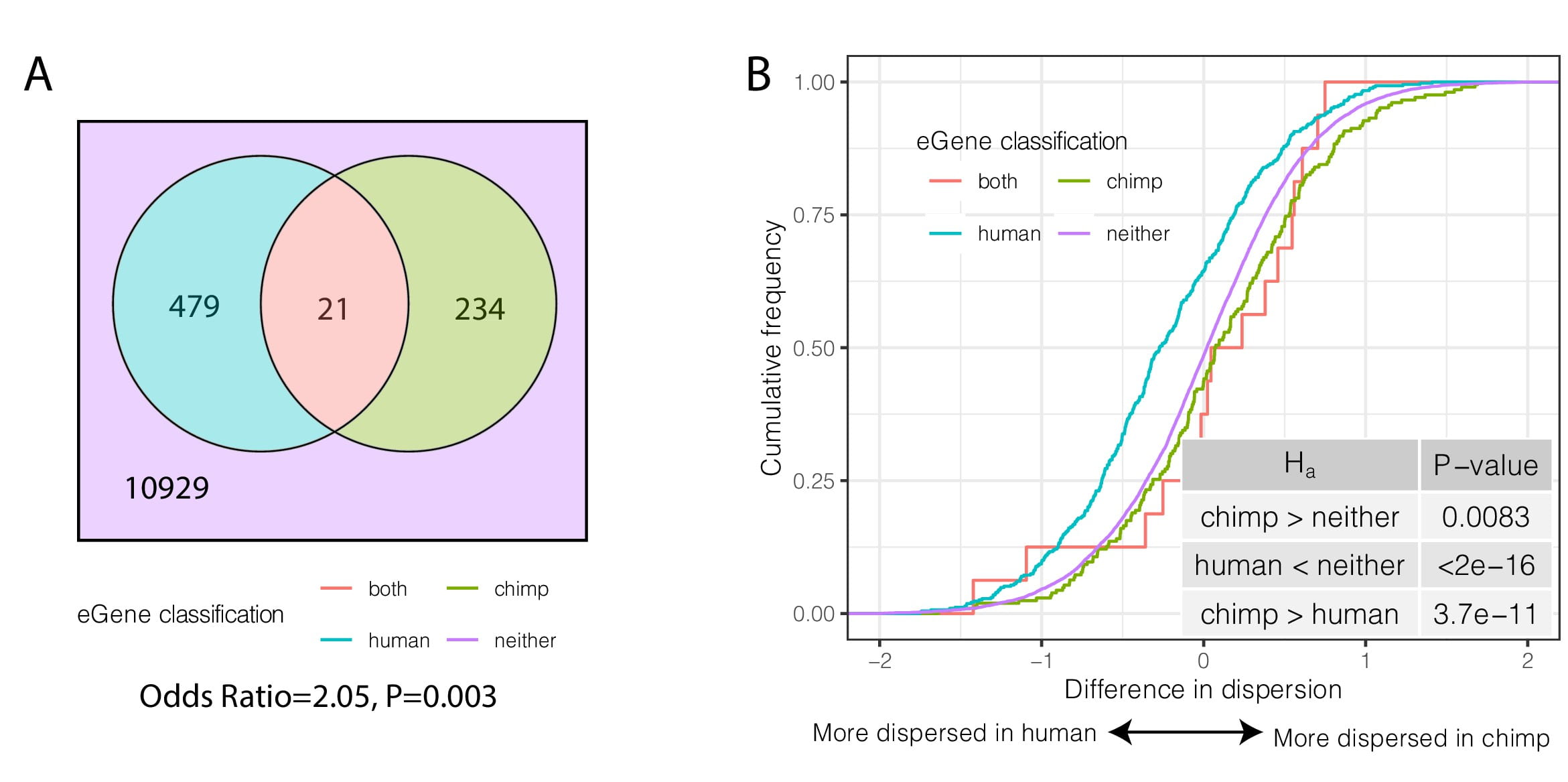
Gene expression variability in human and chimpanzee populations share common determinants
Fair B, Blake LE, Sarkar A, Pavlovic B.J. Cuevas C, and Gilad Y.
eLife; 2020
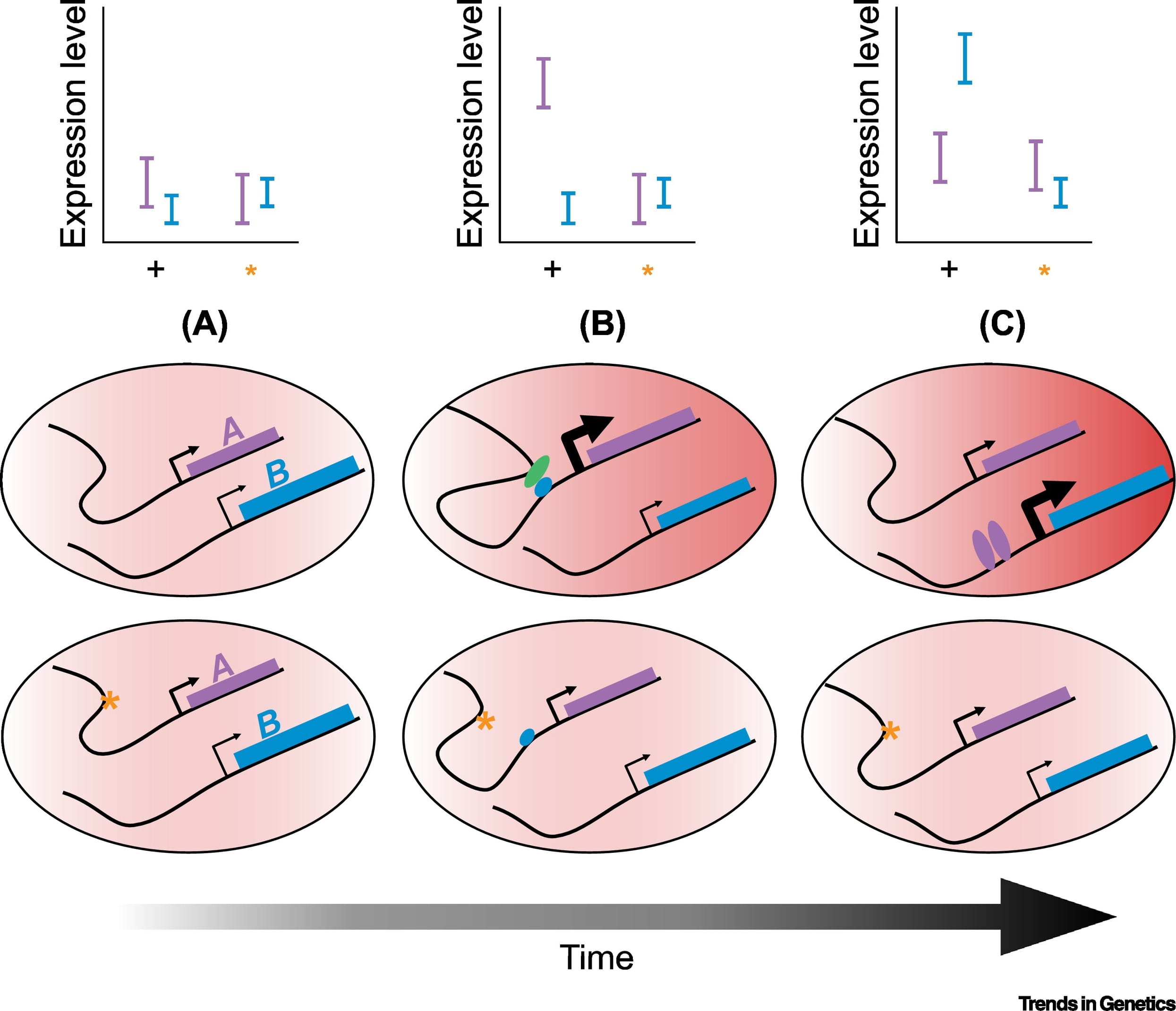
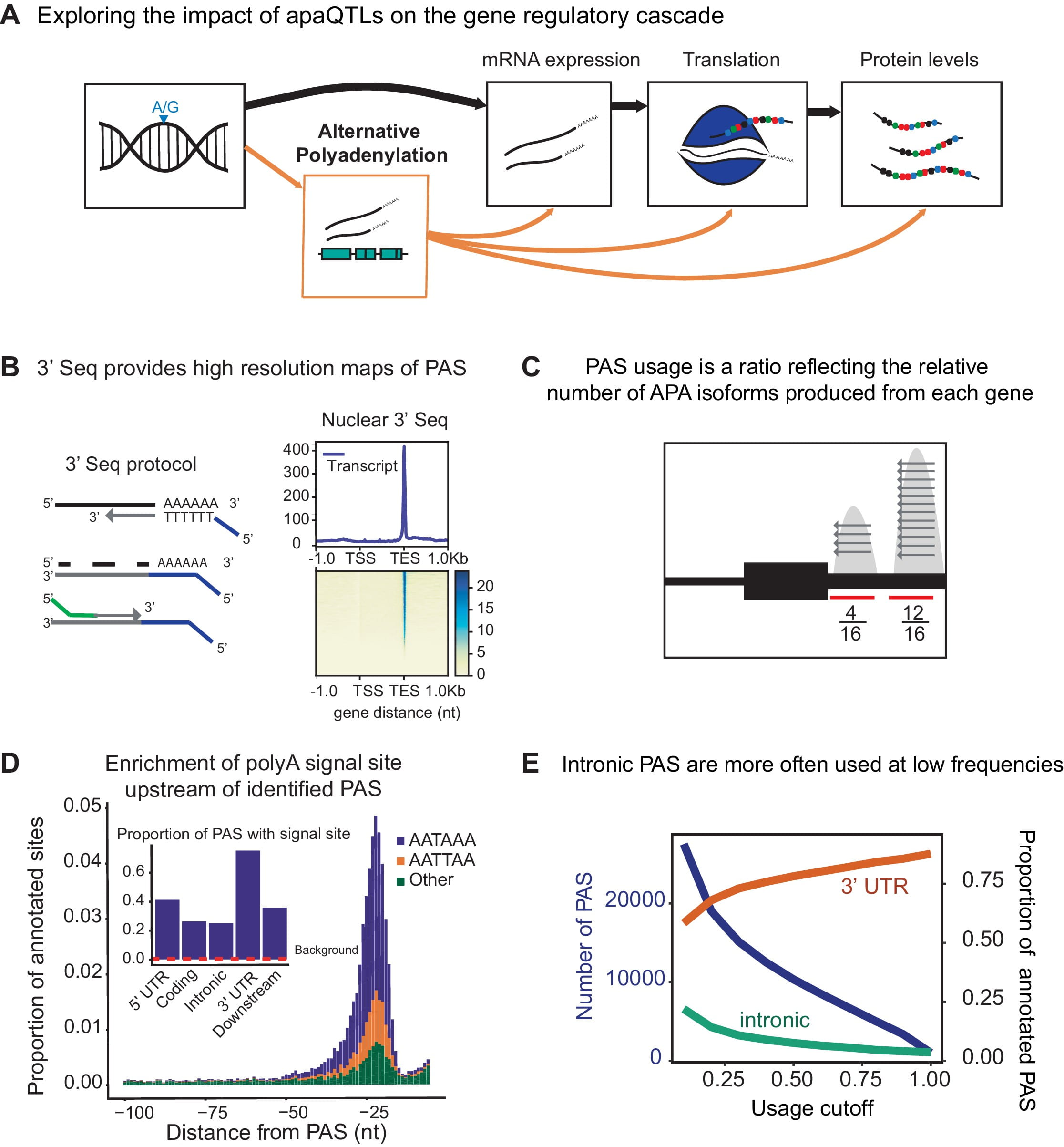
Alternative polyadenylation mediates genetic regulation of gene expression
Mittleman BE, Pott S, Warland S, Zeng T, Mu Z, Kaur M, Gilad Y, and Li YI
eLife; 2020
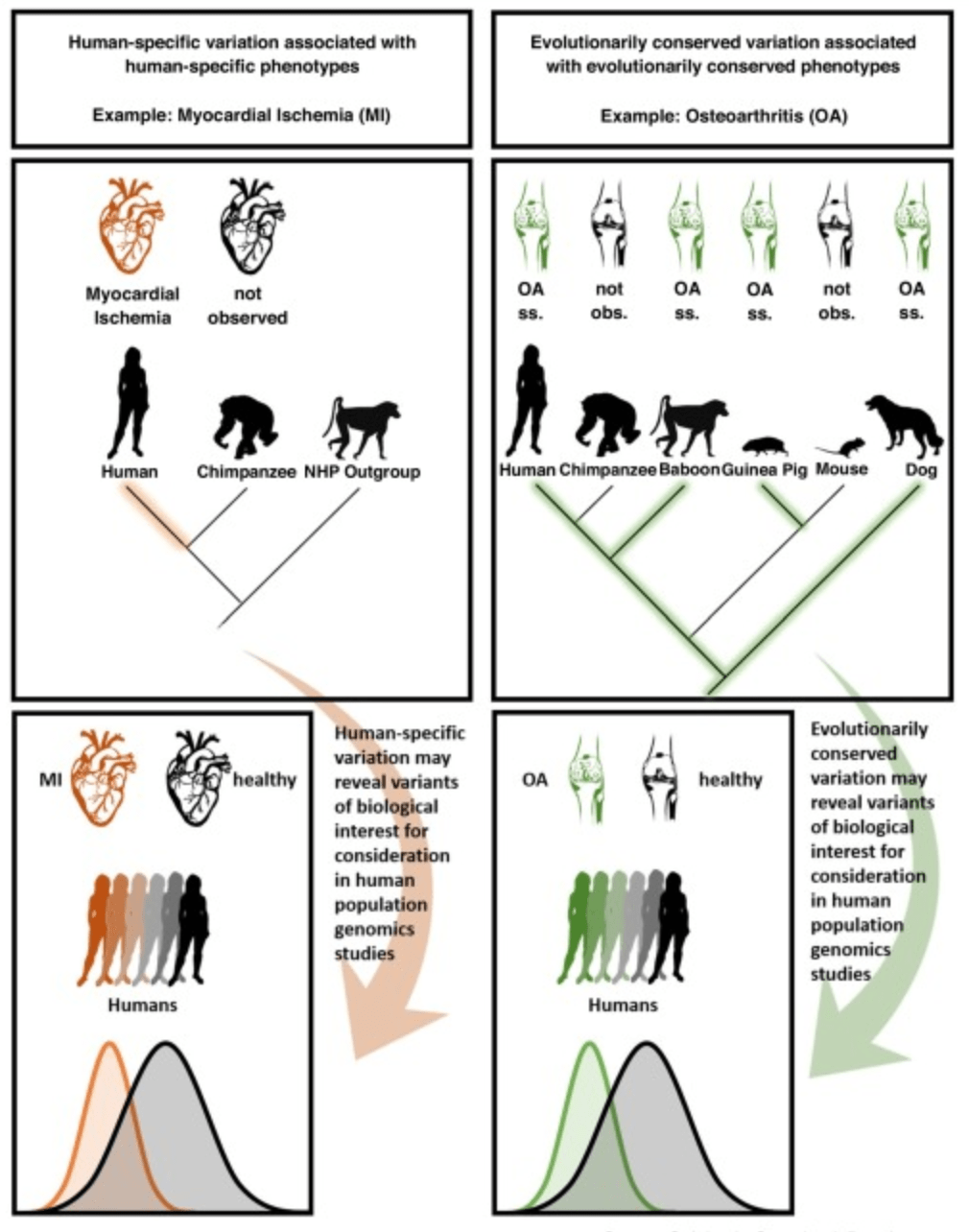
Prime time primate functional genomics
Housman G. and Gilad Y

Effective study design for comparative functional genomics
Kelley JL and Gilad Y
Nature Reviews Genetics; 2020
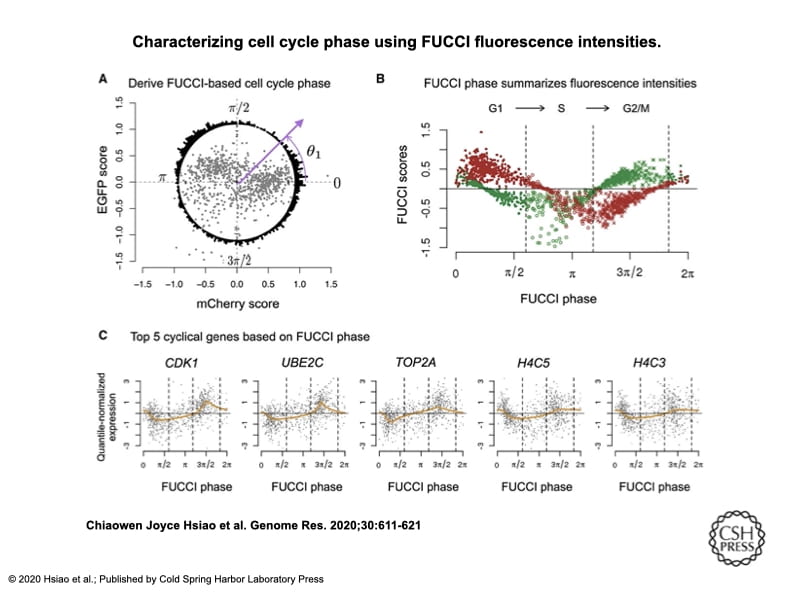
Characterizing and inferring quantitative cell cycle phase in single-cell RNA-seq data analysis
Hsiao CJ, Tung P, Blischak JD, Burnett J, Barr K, Dey KK, Stephens M, and Gilad Y
Genome Research; 2020
Supplementary data: TAR.GZ file
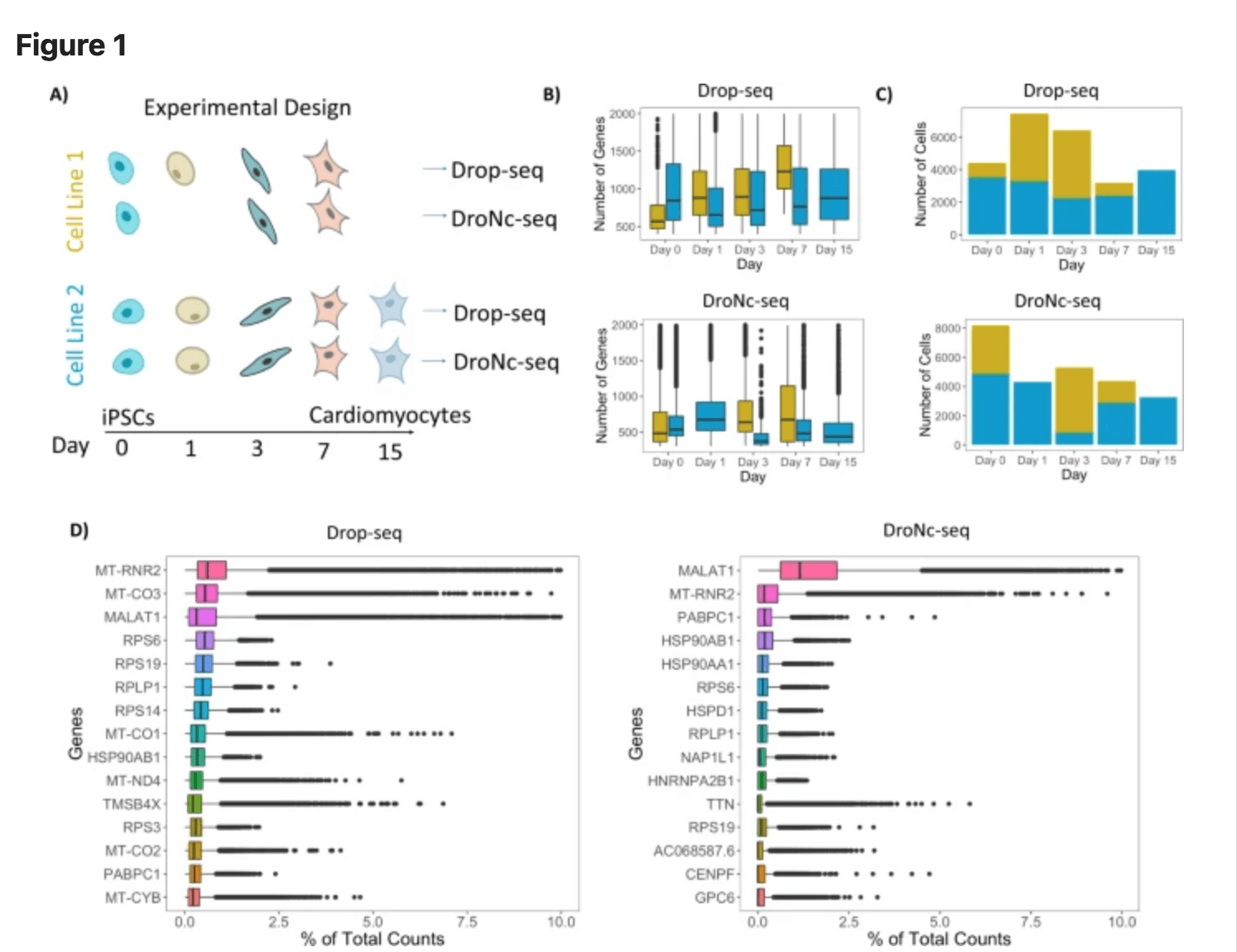
Systematic comparison of high-throughput single-cell and single-nucleus transcriptomes during cardiomyocyte differentiation
Selewa A, Dohn R, Eckart H, Lozano S, Xie B, Gauchat E, Elorbany R, Rhodes K, Burnett J, Gilad Y, Pott S, and Basu A
Scientific Reports; 2020
A comparison of gene expression and DNA methylation patterns across tissues and species
Blake LE, Roux J, Hernando-Herraez I, Banovich NE, Garcia Perez R, Hsiao CJ, Eres I, Chavarria C, Marquez-Bonet T and Gilad YA
Genome Research; 2020
2019
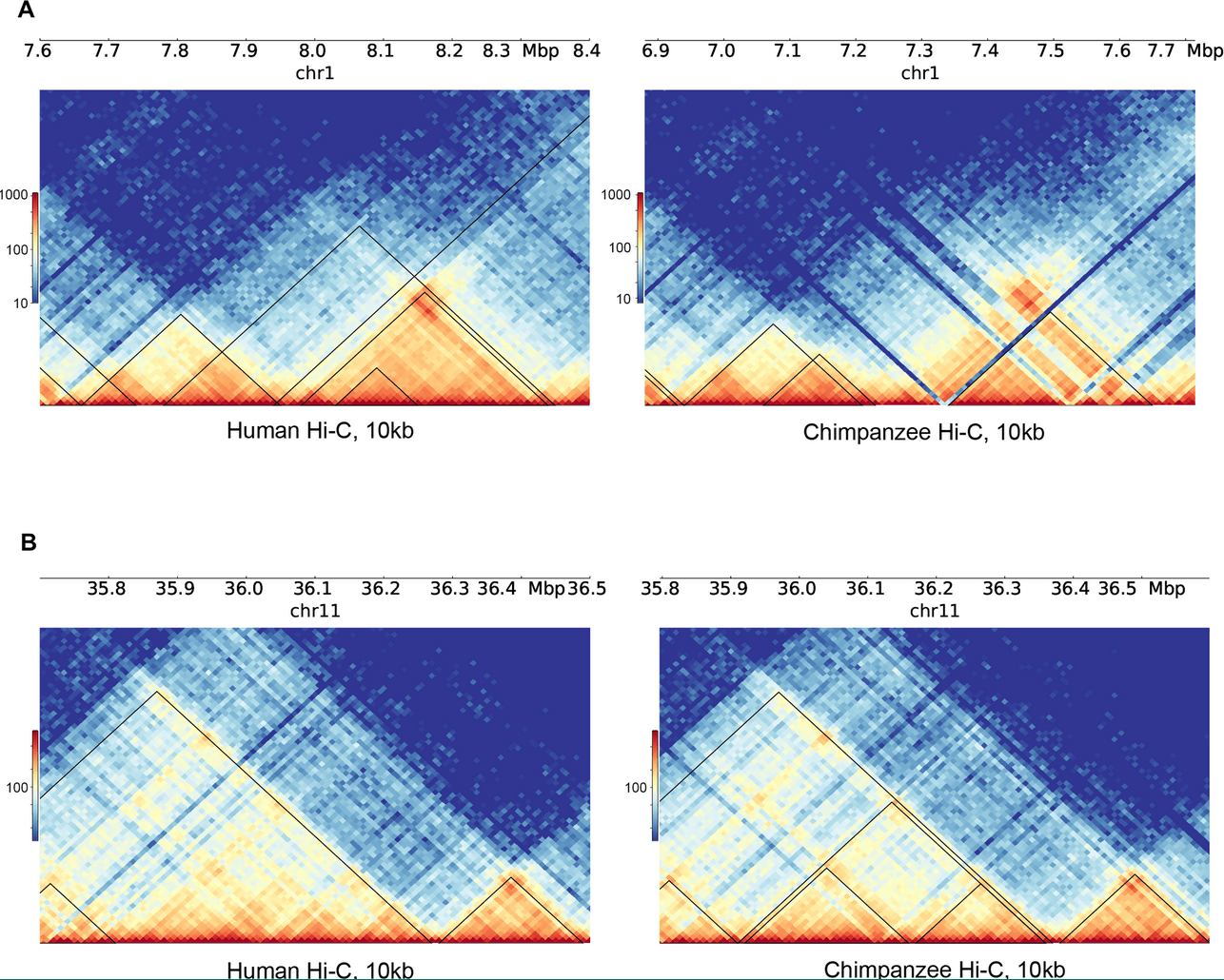
Reorganization of 3D genome structure may contribute to gene regulatory evolution in primates
Eres IE, Luo K, Hsiao CJ, Blake LE and Gilad Y
PLOS Genetics; 2019
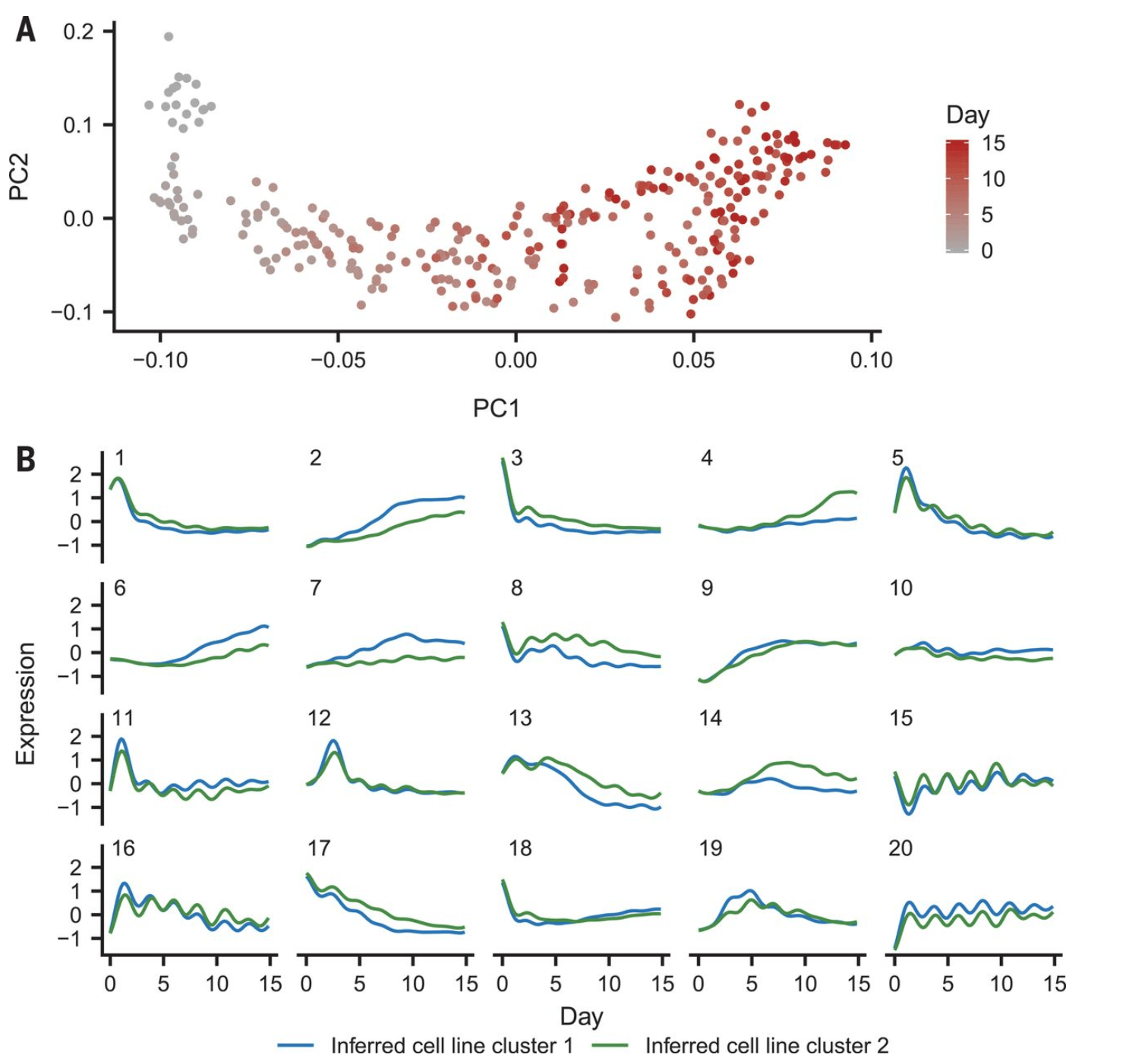
Dynamic genetic regulation of gene expression during cellular differentiation
Strober BJ, Elorbany R, Rhodes K, Krishnan N, Tayeb K, Battle A, and Gilad Y
Science; 2019
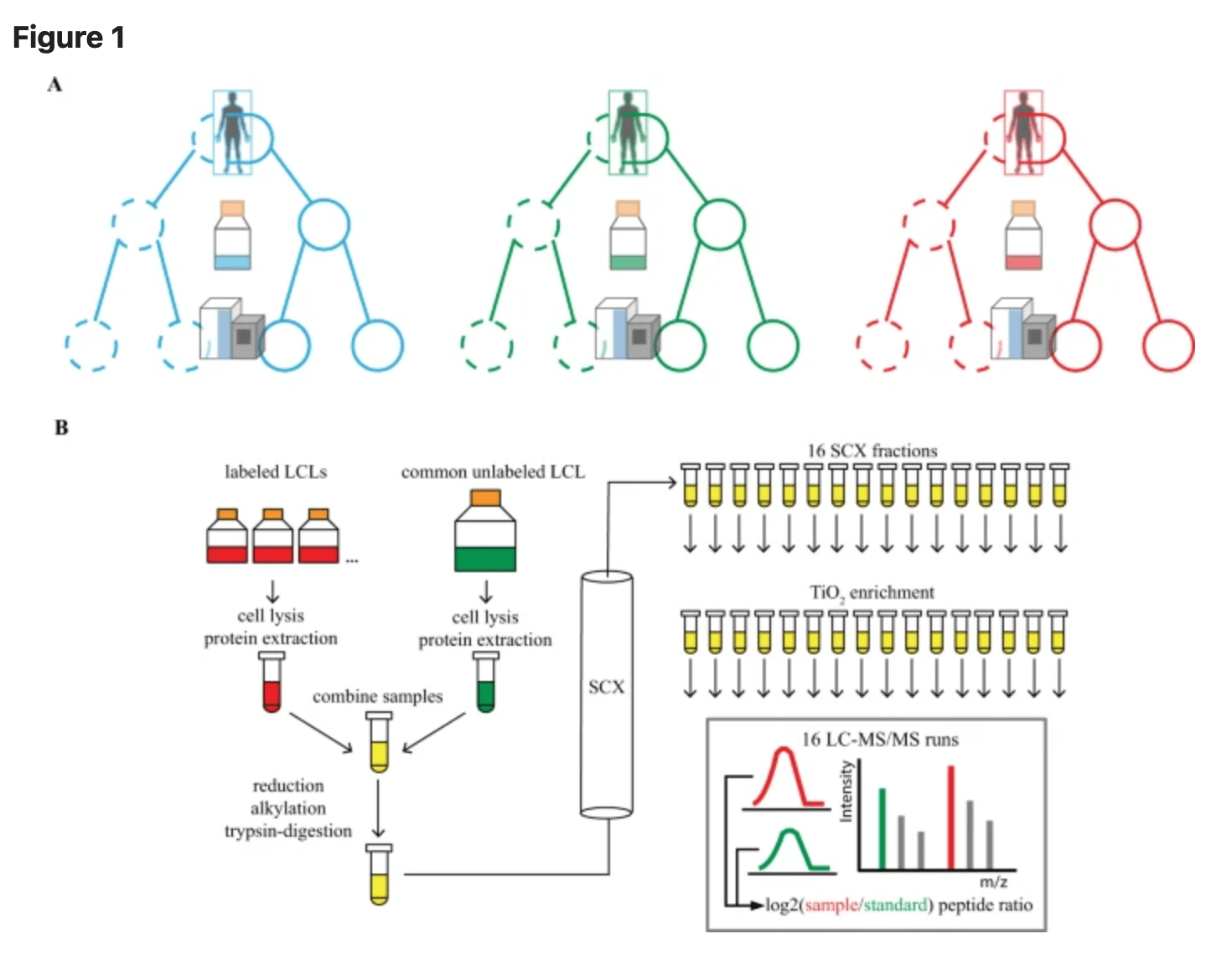
Discovery and characterization of variance QTLs in human induced pluripotent stem cells
Sarkar AK, Tung P, Blischak JD, Burnett JE, Li Yi, Stephens M, and Gilad Y
PLOS Genetics; 2019
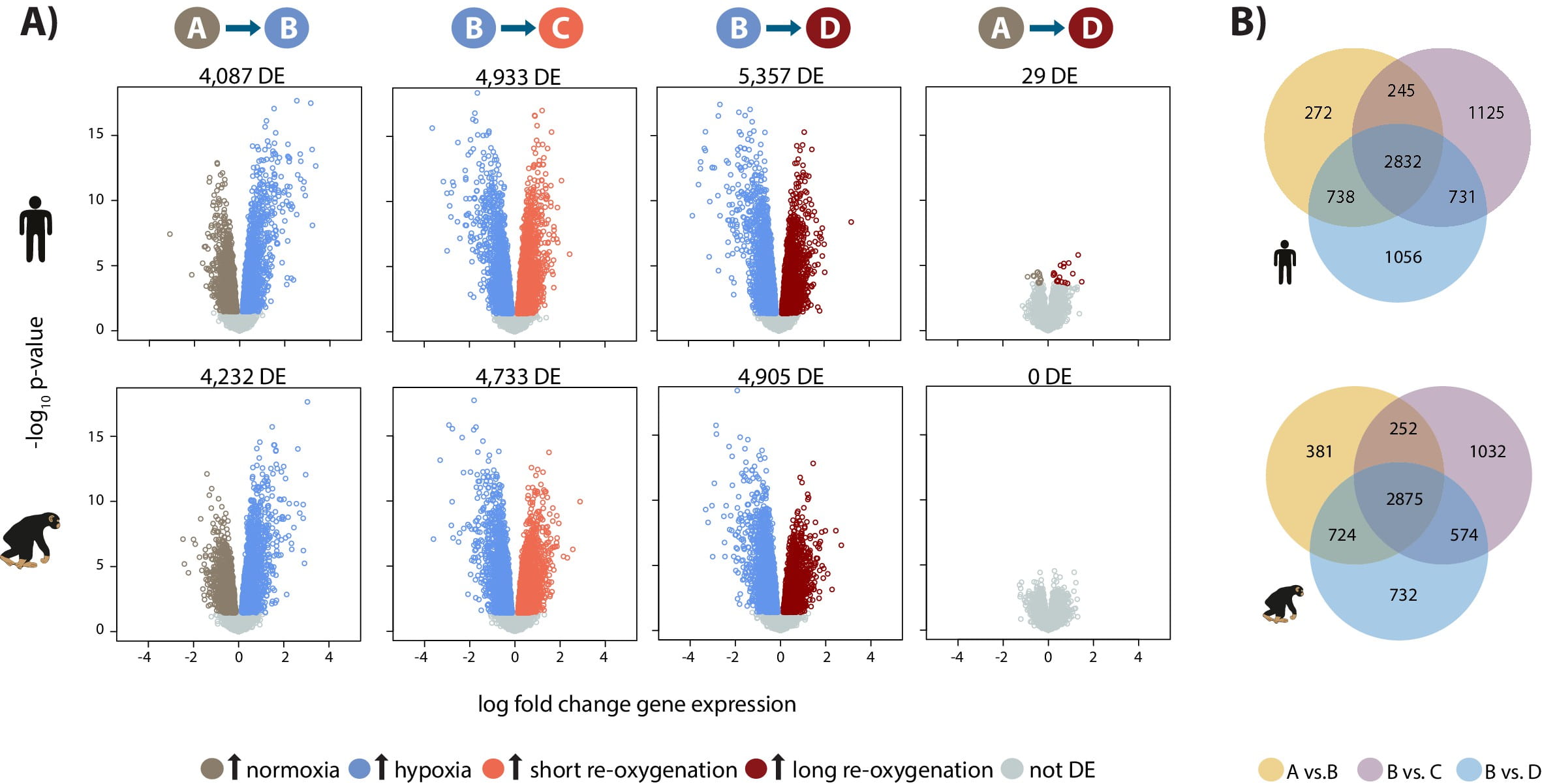
A generally conserved response to hypoxia in iPSC-derived cardiomyocytes from humans and cimpanzees
Ward MC, and Gilad Y
eLife; 2019
2018

A comparative study of endoderm differentiation in humans and chimpanzees
Blake LE, Thomas SJ, Blischak JD, Hsiao C, Chavarria C, Myrthil M, Gilad Y, Pavlovic BJ
Genome Biology; 2018
Supplementary data: XLS file

A Comparative Assessment of Human and Chimpanzee iPSC-derived Cardiomyocytes with Primary Heart Tissues
Pavlovic BJ, Blake LE, Roux J, Chavarria C, and Gilad Y
Scientific Reports; 2018
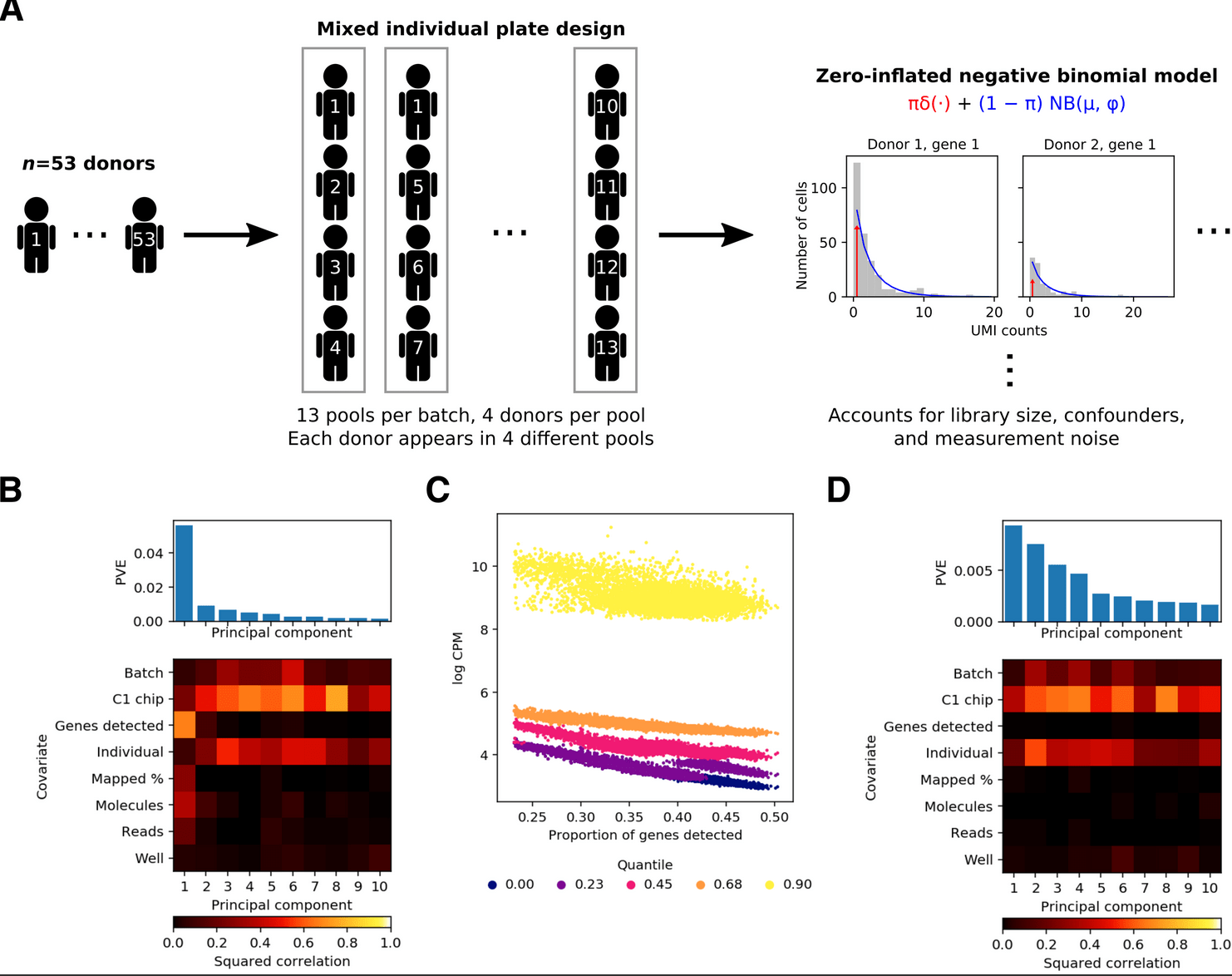
A Methodological Assessment and Characterization of Genetically-Driven Variation in Three Human Phosphoproteomes
Engelmann BW, Hsiao CJ, Blischak JD, Fourne Y, Ford M, and Gilad Y
Scientific Reports; 2018
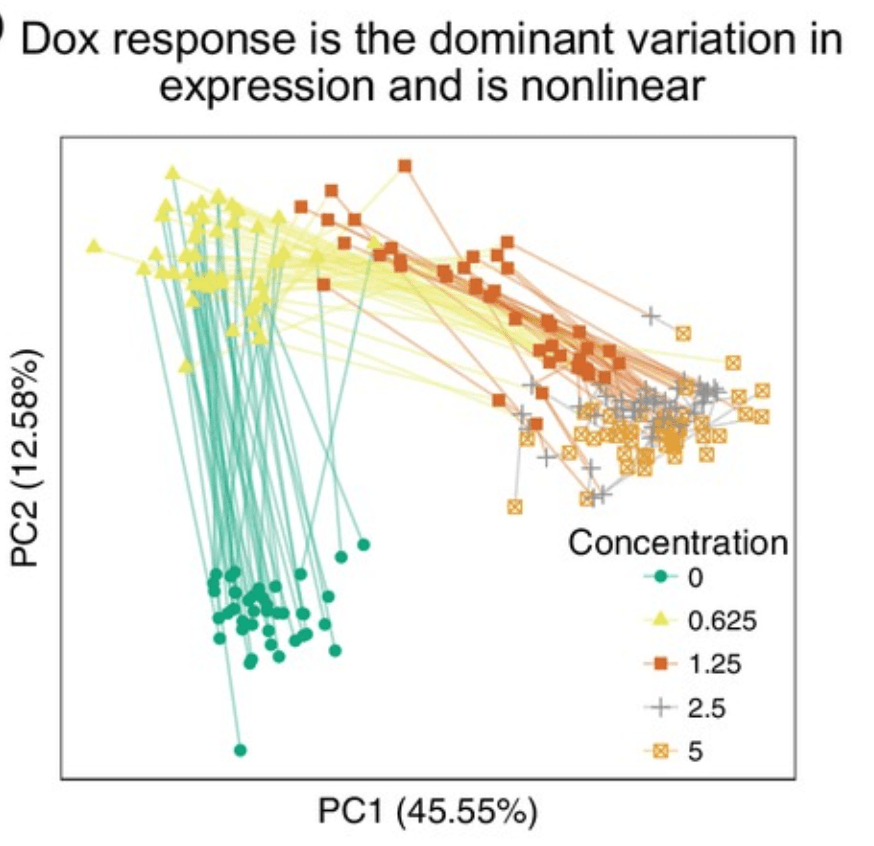
Determining the genetic basis of anthracycline-cardiotoxicity by molecular response QTL mapping in induced cardiomyocytes
Knowles DA, Burrows CK, Blischak JD, Patterson KM, Ober C, Pritchard JK, and Gilad Y
eLife; 2018.

Silencing of Transposable Elements May Not Be A Major Driver Of Regulatory Evolution in Primate iPSCs
Ward MC, Zhao S, Luo K, Pavlovic BJ, Karimi MM, Stephens M, and Gilad Y
eLife; 2018
2017
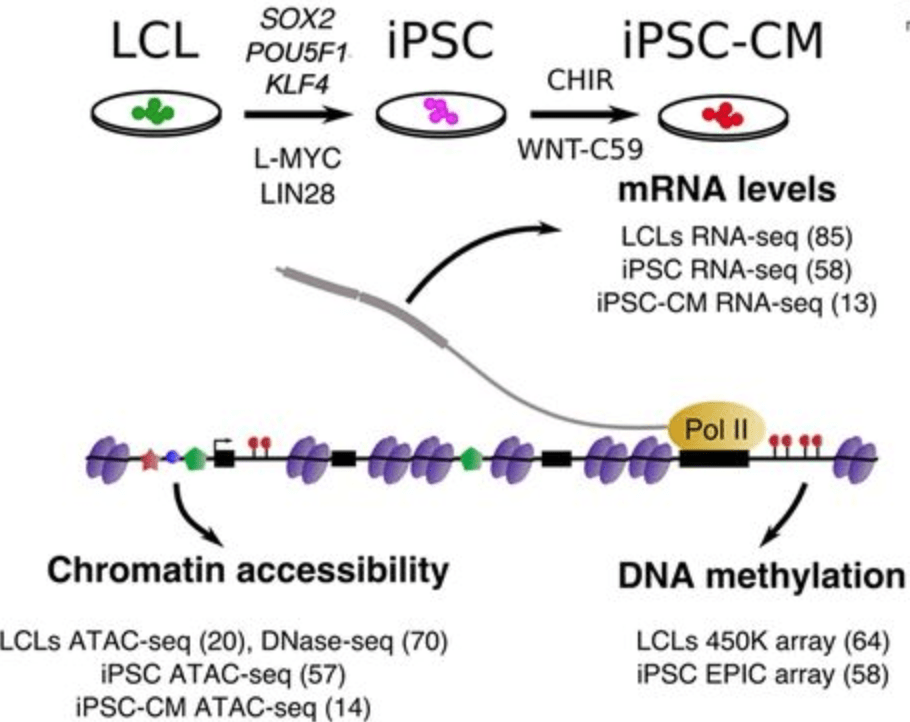
Impact of regulatory variation across human iPSCs and differentiated cells
Banovich N, Li YI, Raj A, Ward MC, Greenside P, Calderon D, Tung P, Burnett JE, Myrthil M, Thomas SM, Burrows CK, Gallego Romero I, Pavlovic BJ, Kundaje A, Pritchard JK, and Gilad Y
Genome Research; 2017
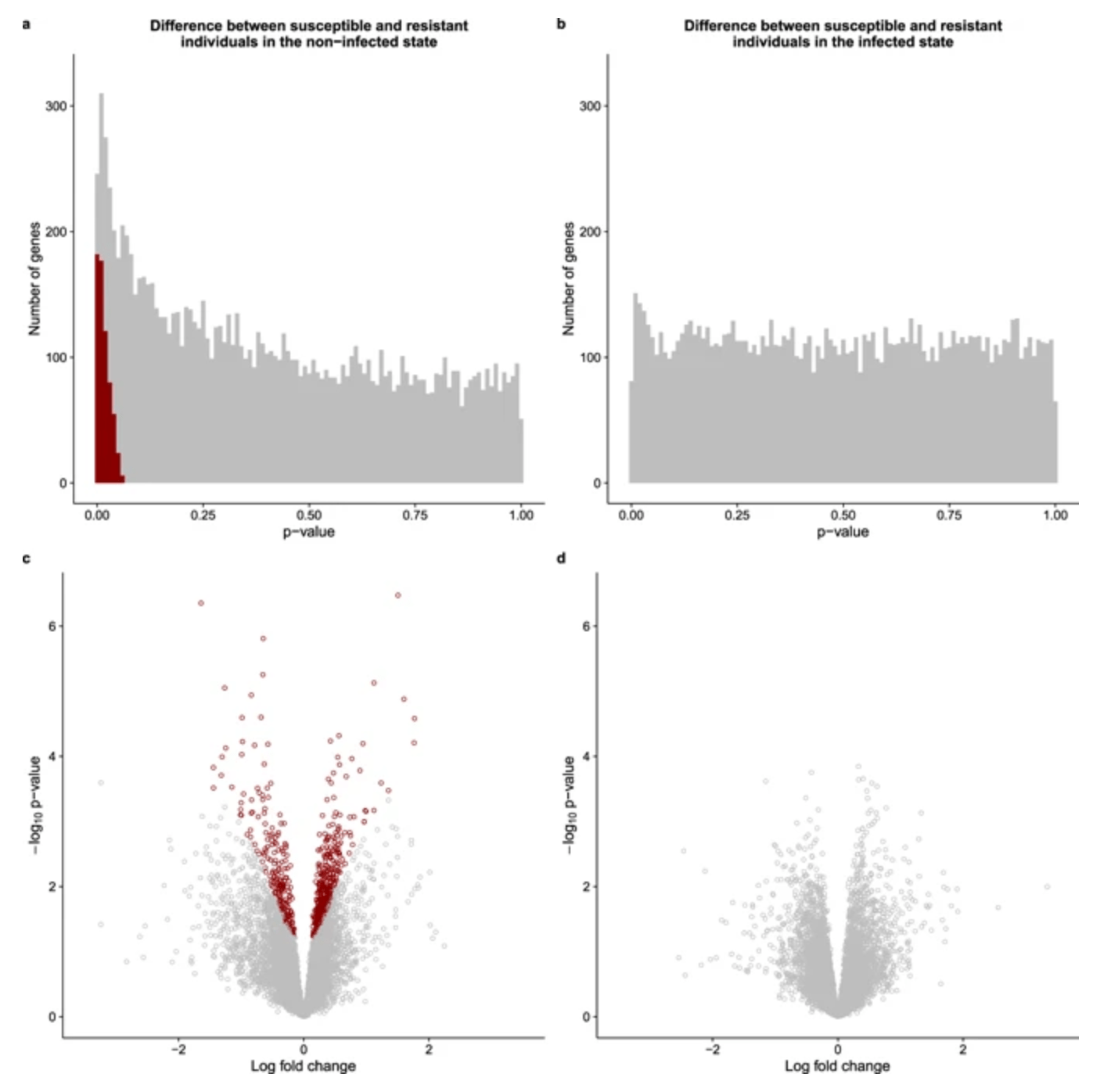
Predicting susceptibility to tuberculosis based on gene expression profiling in dendritic cells
Blischak JD, Tailleux L, Myrthil M, Charlois C, Bergot E, Dinh A, Morizot G, Chény O, Platen CV, Herrmann J, Brosch R, Barreiro LB, and Gilad Y
Scientific Reports; 2017
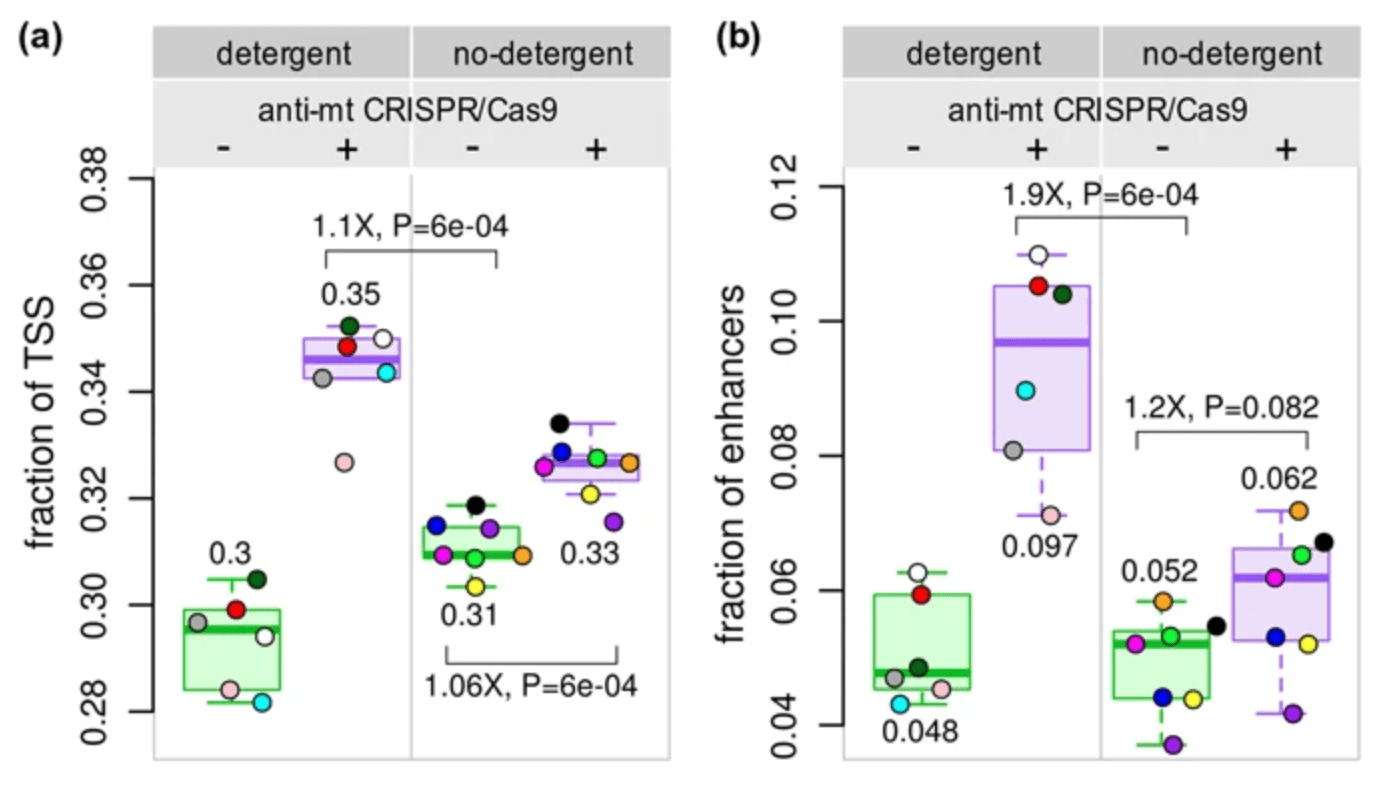
Reducing mitochondrial reads in ATAC-seq using CRISPR/Cas9
Montefiori L, Hernandez L, Zhang Z, Gilad Y, Ober C, Crawford G, Nobrega M, and Jo Sakabe N
Scientific Reports; 2017
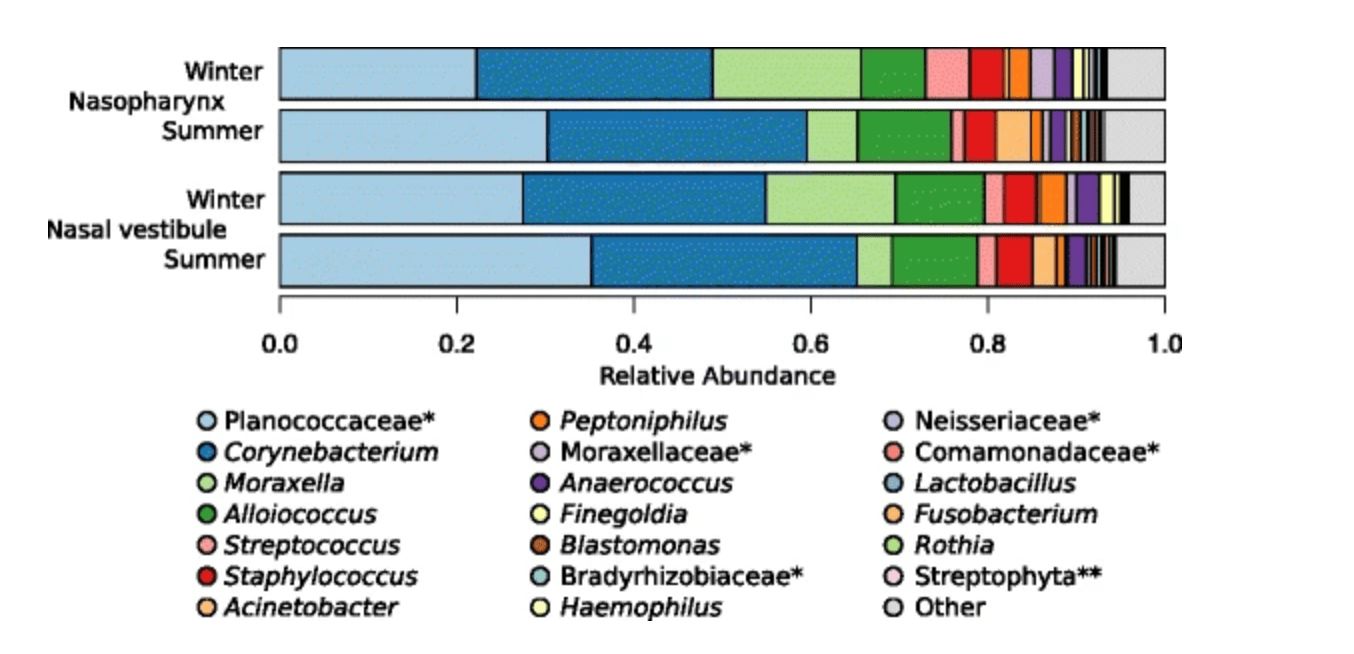
Host genetic variation in mucosal immunity pathways influences the upper airway microbiome
Igartua C, Davenport ER, Gilad Y, Nicolae DL, Pinto J, and Ober C
Microbiome; 2017
Batch effects and the effective design of single-cell gene expression studies
Tung P, Blischak JD, Hsiao C, Knowles DA, Burnett JE, Pritchard JK, and Gilad Y
Scientific Reports; 2017
2016
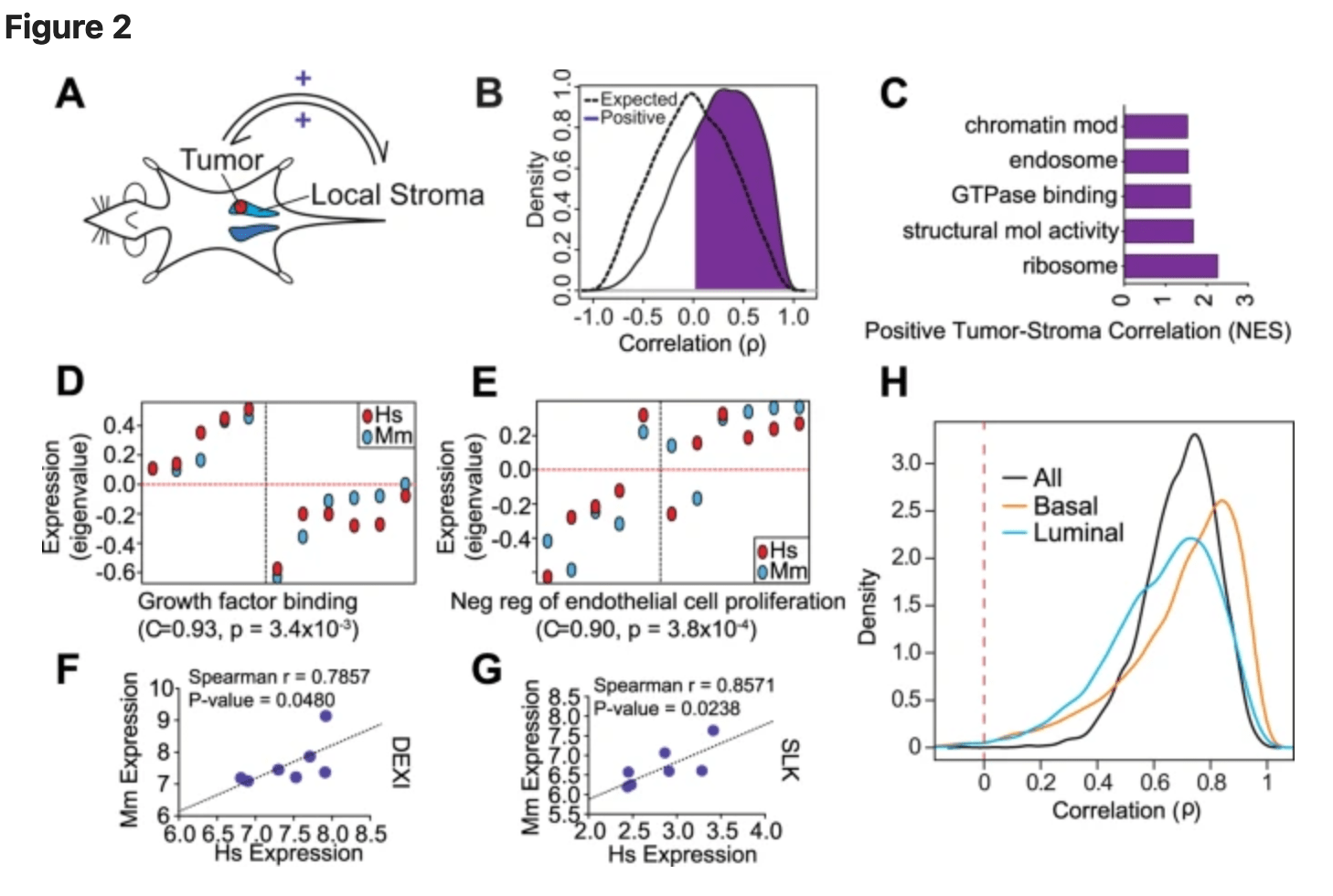
Gene expression in local stroma reflects breast tumor states and predicts patient outcome
Bainer R, Frankenberger C, Rabe D, An G, Gilad Y, and Rosner MR
Scientific Reports; 2016
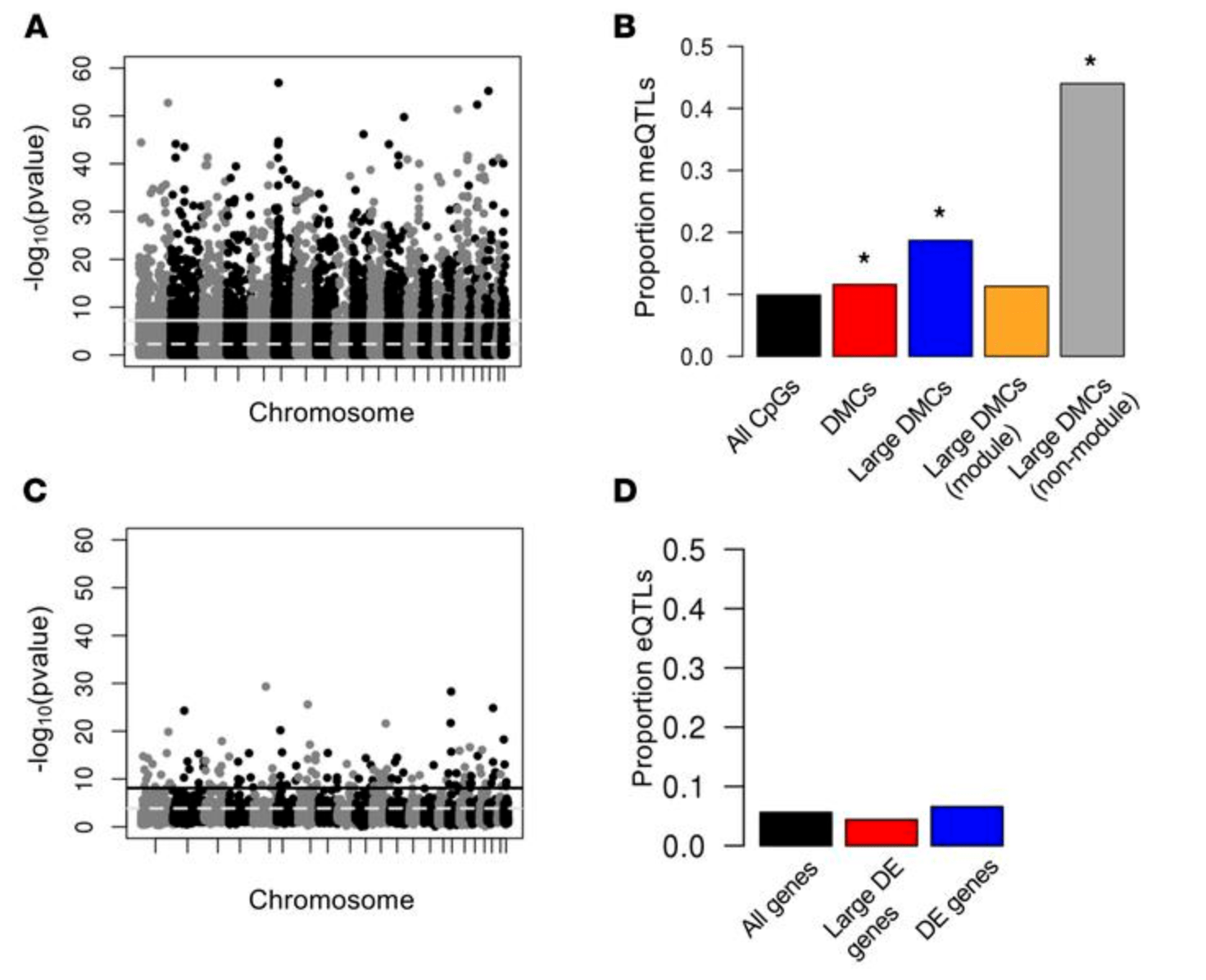
DNA methylation in lung cells is associated with asthma endotypes and genetic risk
Nicodemus-Johnson J, Myers RA, Sakabe NJ, Sobreira DR, Hogarth DK, Naureckas ET, Sperling AI, Solway J, White SR, Nobrega MA, Nicolae DL, Gilad Y, and Ober C
JCI Insight; 2016
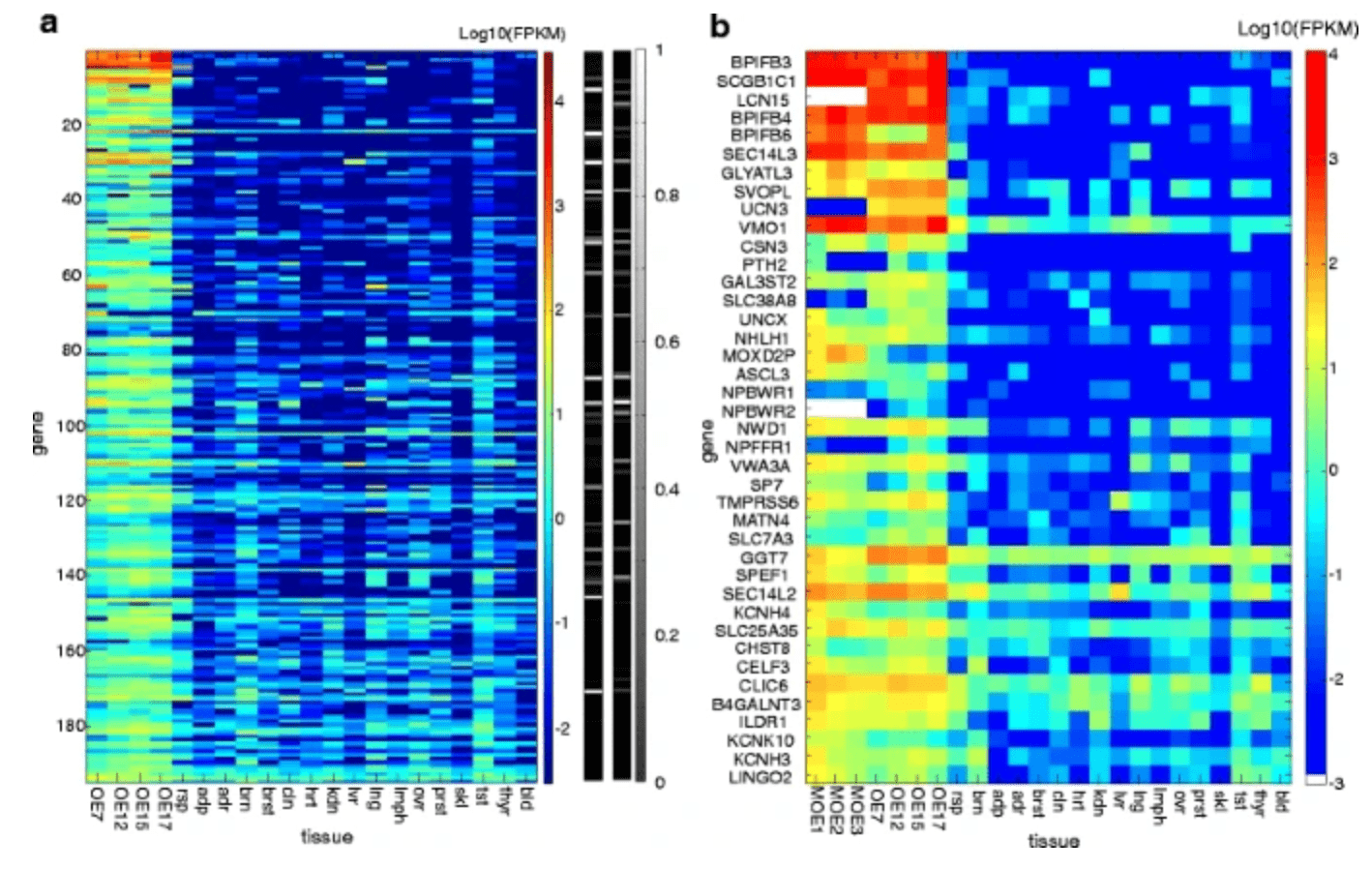
The human olfactory transcriptome
Olender T, Keydar I, Pinto JM, Tatarskyy P, Alkelai A, Chien MS, Fishilevich S, Restrepo D, Matsunami H, Gilad Y, and Lancet D
BMC Genomics; 2016

Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling
Raj A, Wang SH, Shim H, Harpak A, Li YI, Engelmann B, Stephens M, Gilad Y, and Pritchard JK
eLife; 2016
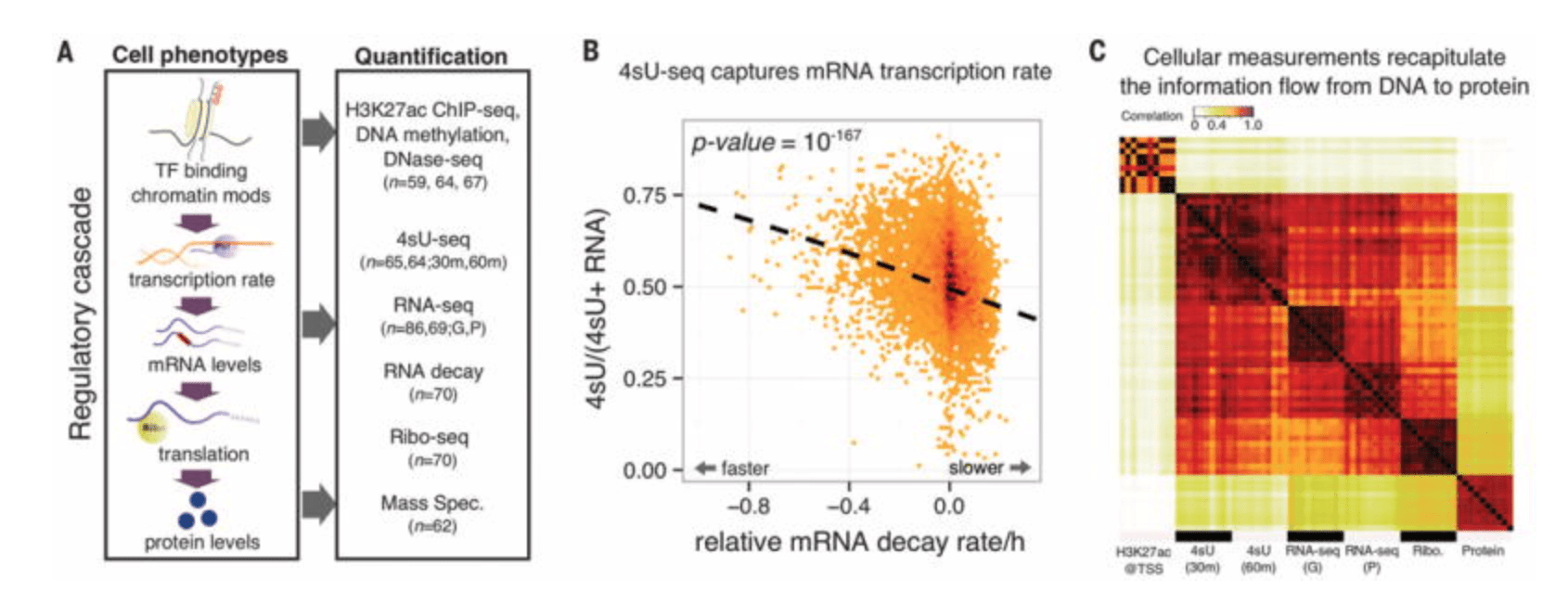
RNA splicing is a primary link between genetic variation and diseases
Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, and Pritchard JK
Science; 2016
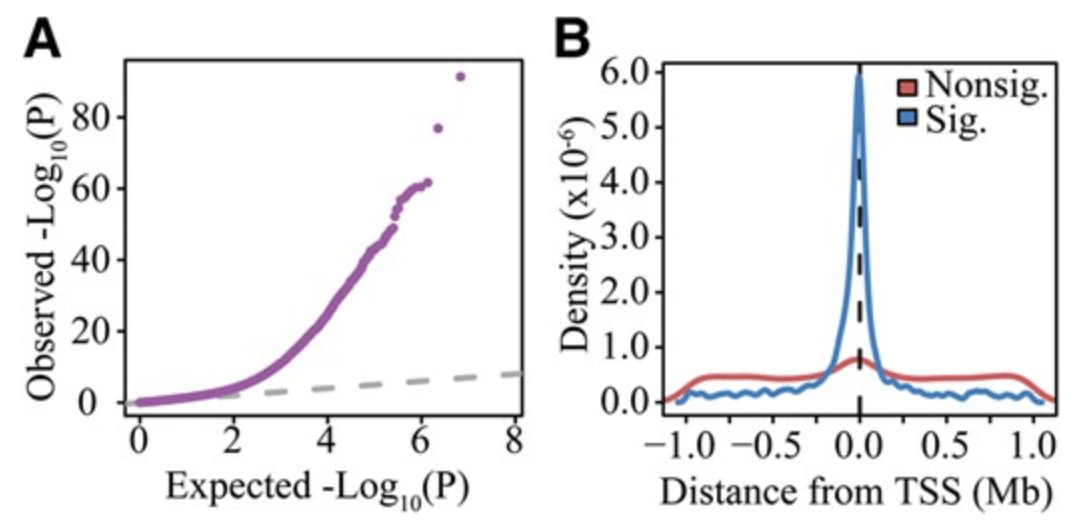
Integrated Analyses of Gene Expression and Genetic Association Studies in a Founder Population
Cusanovich DA, Caliskan M, Billstrand C, Michelini K, Chavarria C, De Leon S, Mitrano A, Lewellyn N, Elias JA, Chupp GL, Lang RM, Shah SJ, DeCara JM, Gilad Y, and Ober C
Human Molecular Genetics; 2016
Supplementary data: ZIP file
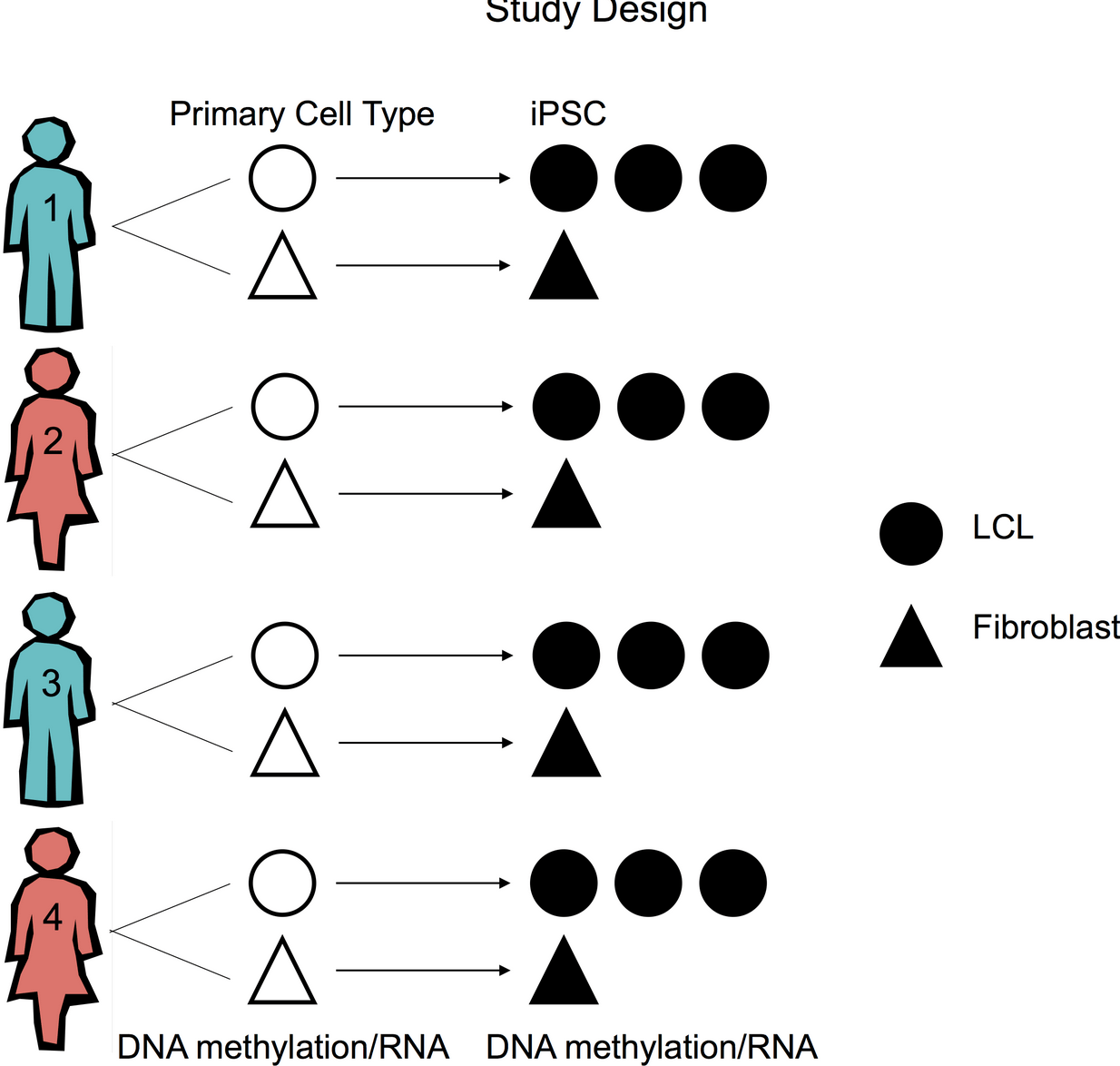
Genetic Variation, Not Cell Type of Origin, Underlies the Majority of Identifiable Regulatory Differences in iPSCs
Burrows CK, Banovich NE, Pavlovic BJ, Patterson K, Gallego Romero I, Pritchard JK, and Gilad Y
PLoS Genetics; 2016
Supplementary data: ZIP file
2015
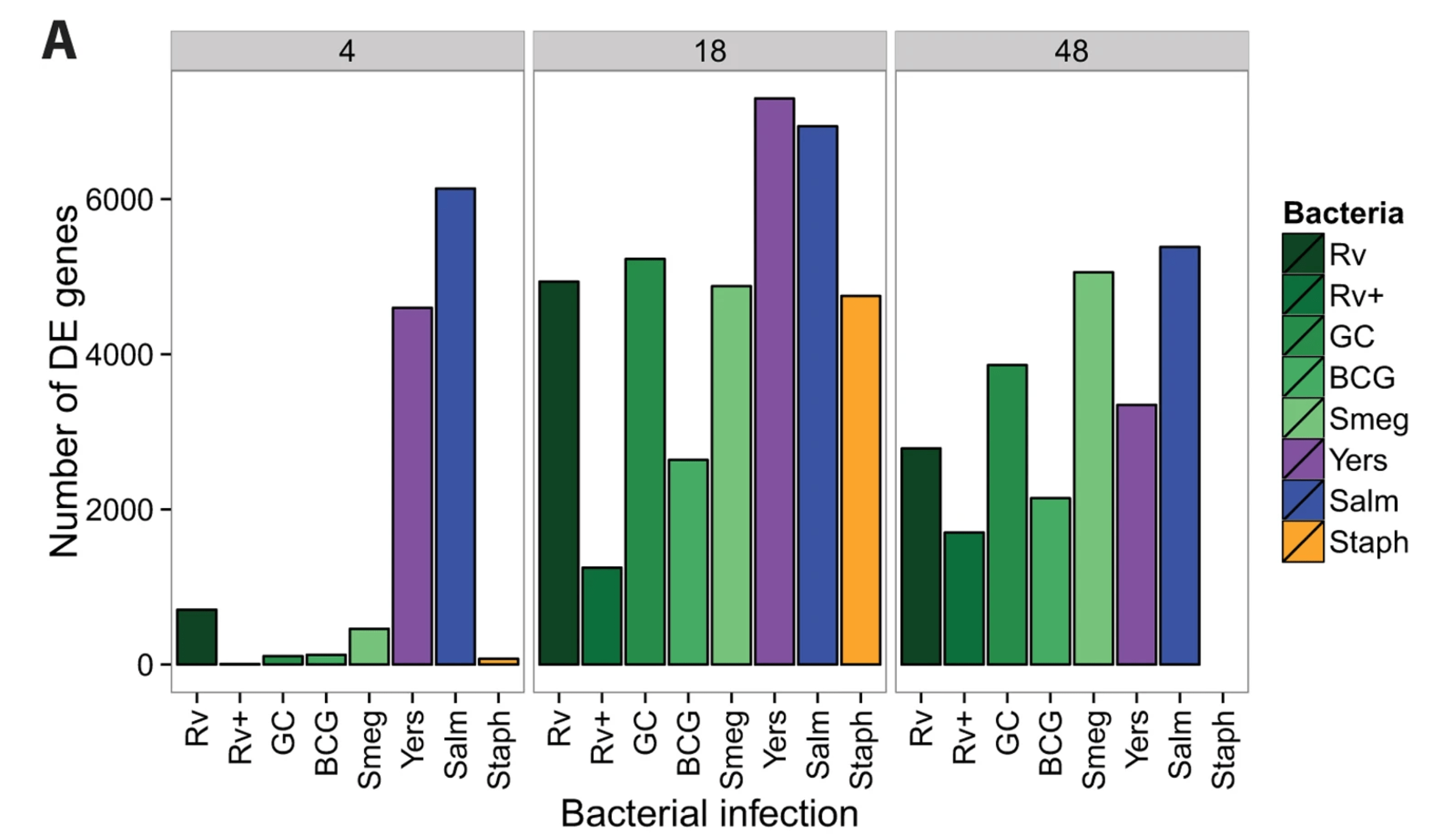
Mycobacterial infection induces a specific human innate immune response
Blischak JD, Tailleux L, Mitrano A, Barreiro L, and Gilad Y
Scientific Reports; 2015
Supplementary data: ZIP file
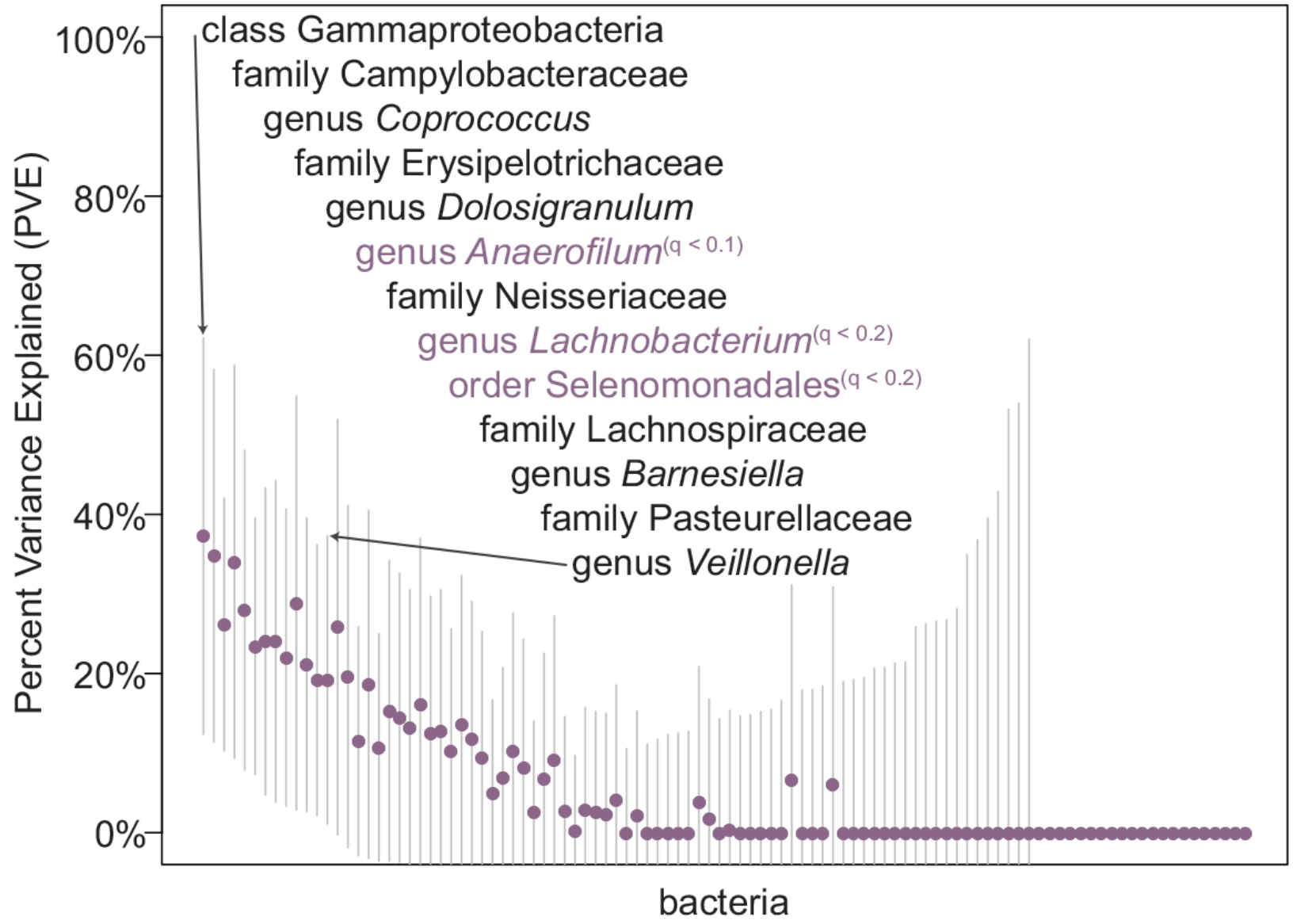
Genome-Wide Association Studies of the Human Gut Microbiota
Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, and Gilad Y
PLoS One; 2015
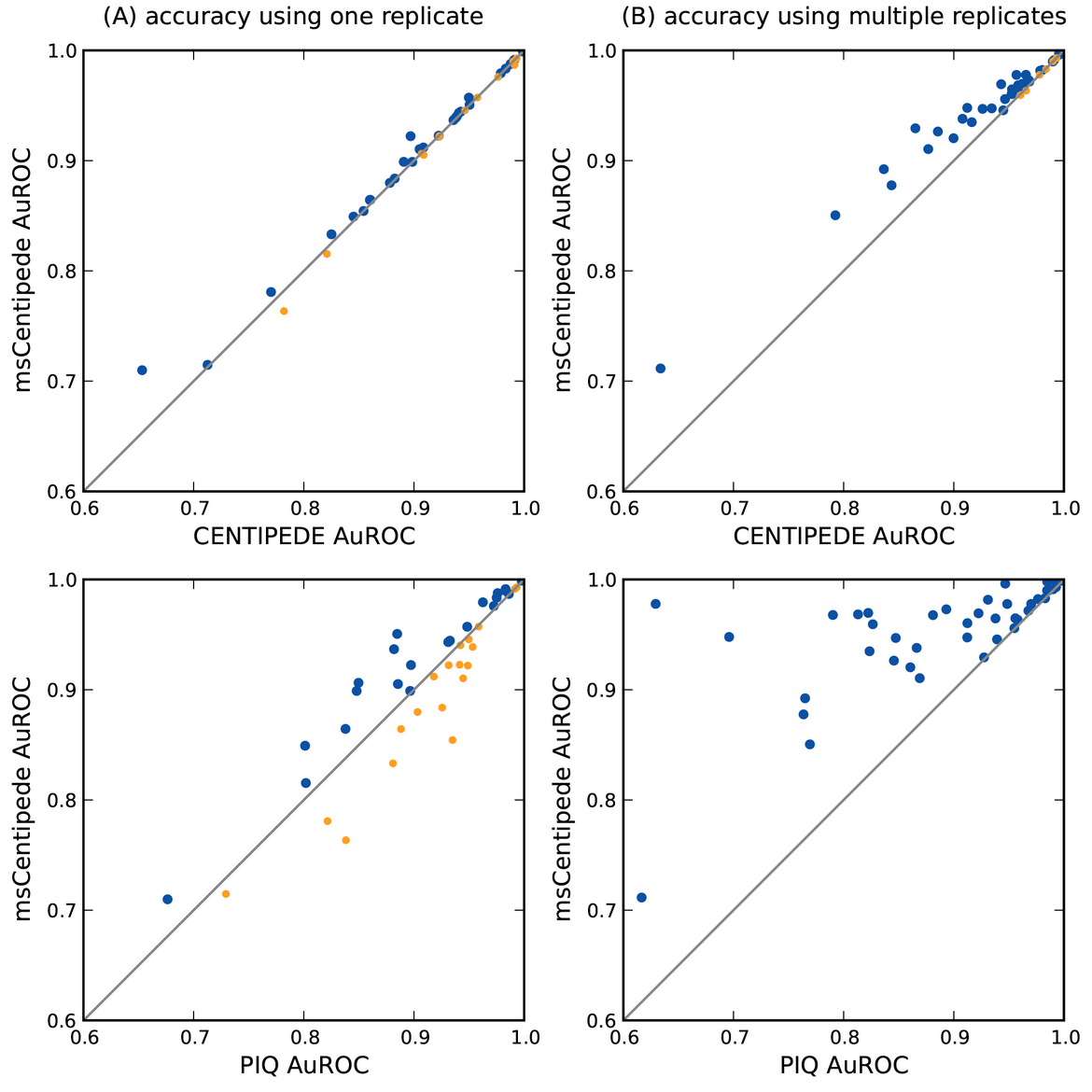
msCentipede: Modeling Heterogeneity across Genomic Sites and Replicates Improves Accuracy in the Inference of Transcription Factor Binding
Raj A, Shim H, Gilad Y, Pritchard JK, and Stephens M
PLoS One; 2015
Bacterial infection remodels the DNA methylation landscape of human dendritic cells
Pacis A, Tailleux L, Morin AM, Lambourne J, Maclsaac JL, Yotova V, Dumaine A, Danckaert A, Luca F, Grenier JC, Hansen KD, Gicquel B, Yu M, Pai A, He C, Tung J, Pastinen T, Kobor MS, Pique-Regi R, Gilad Y, and Barreiro LB
Genome Research; 2015
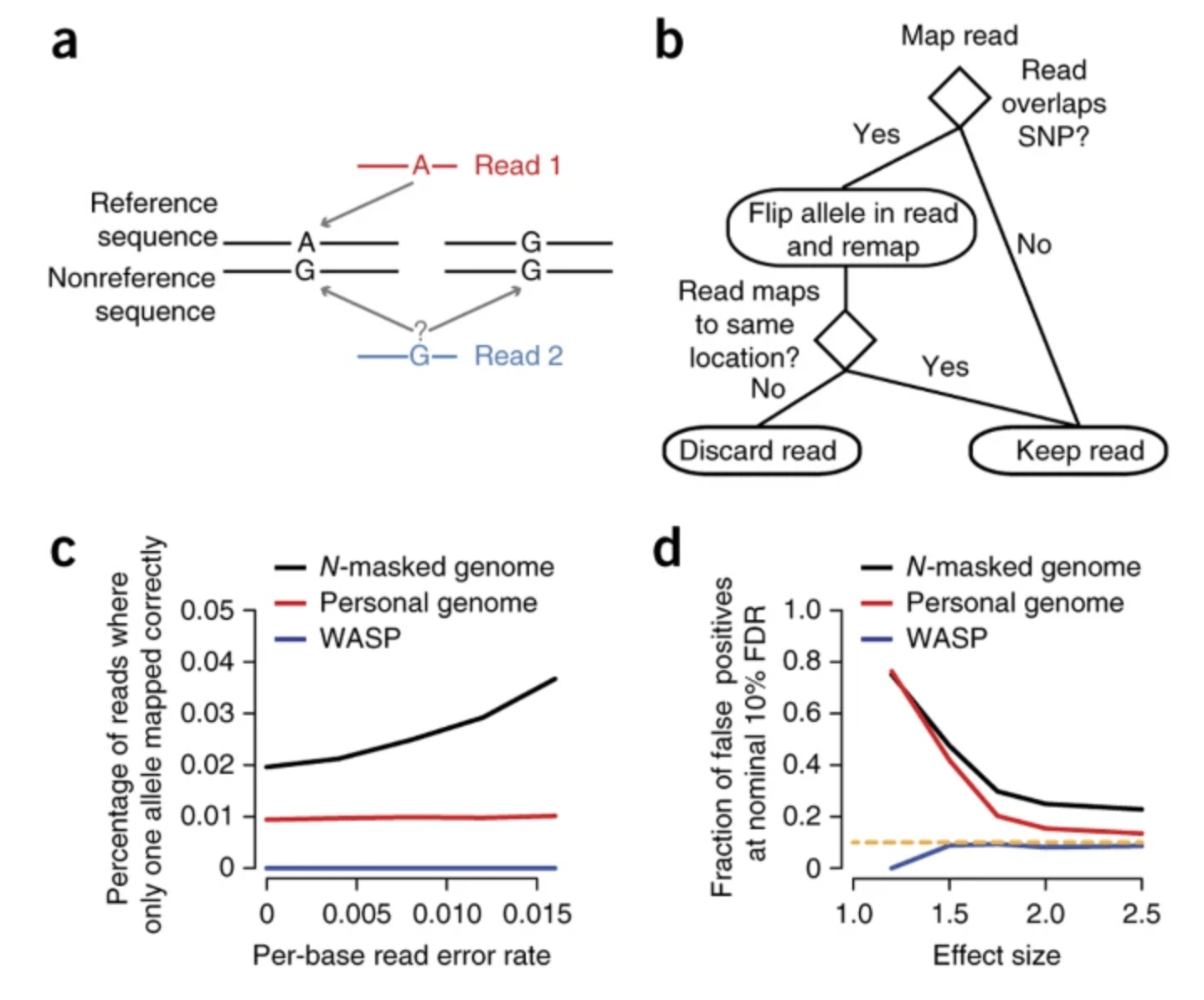
WASP: allele-specific software for robust molecular quantitative trait locus discovery
van de Geijn B, McVicker G, Gilad Y, Pritchard JK
Nature Methods; 2015
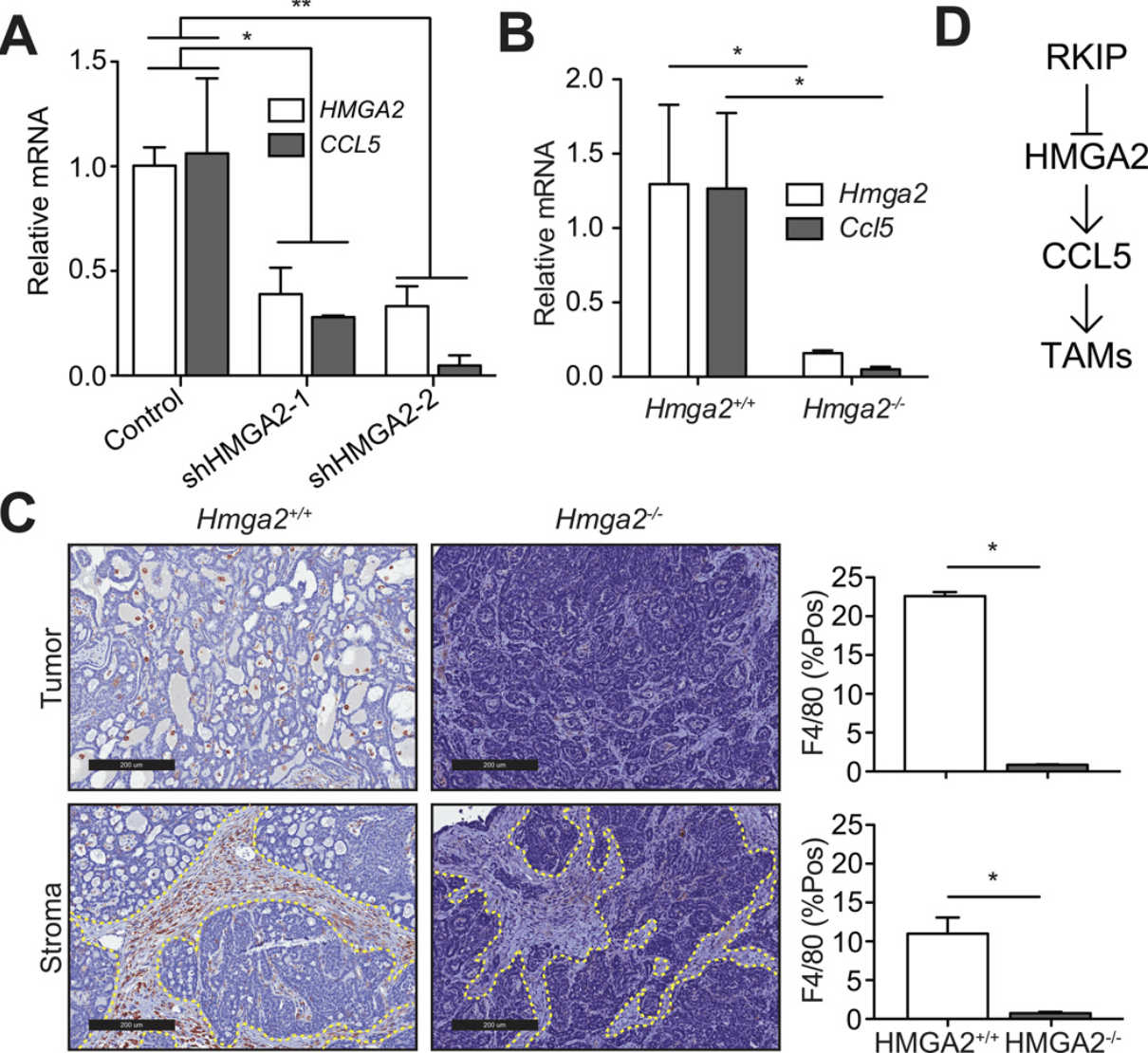
Metastasis suppressors regulate the tumor microenvironment by blocking recruitment of pro-metastatic tumor-associated macrophages
Frankenberger C, Rabe D, Bainer R, Sankarasharma D, Chada K, Krausz T, Gilad Y, Becker L, and Rosner MR
Cancer Research; 2015
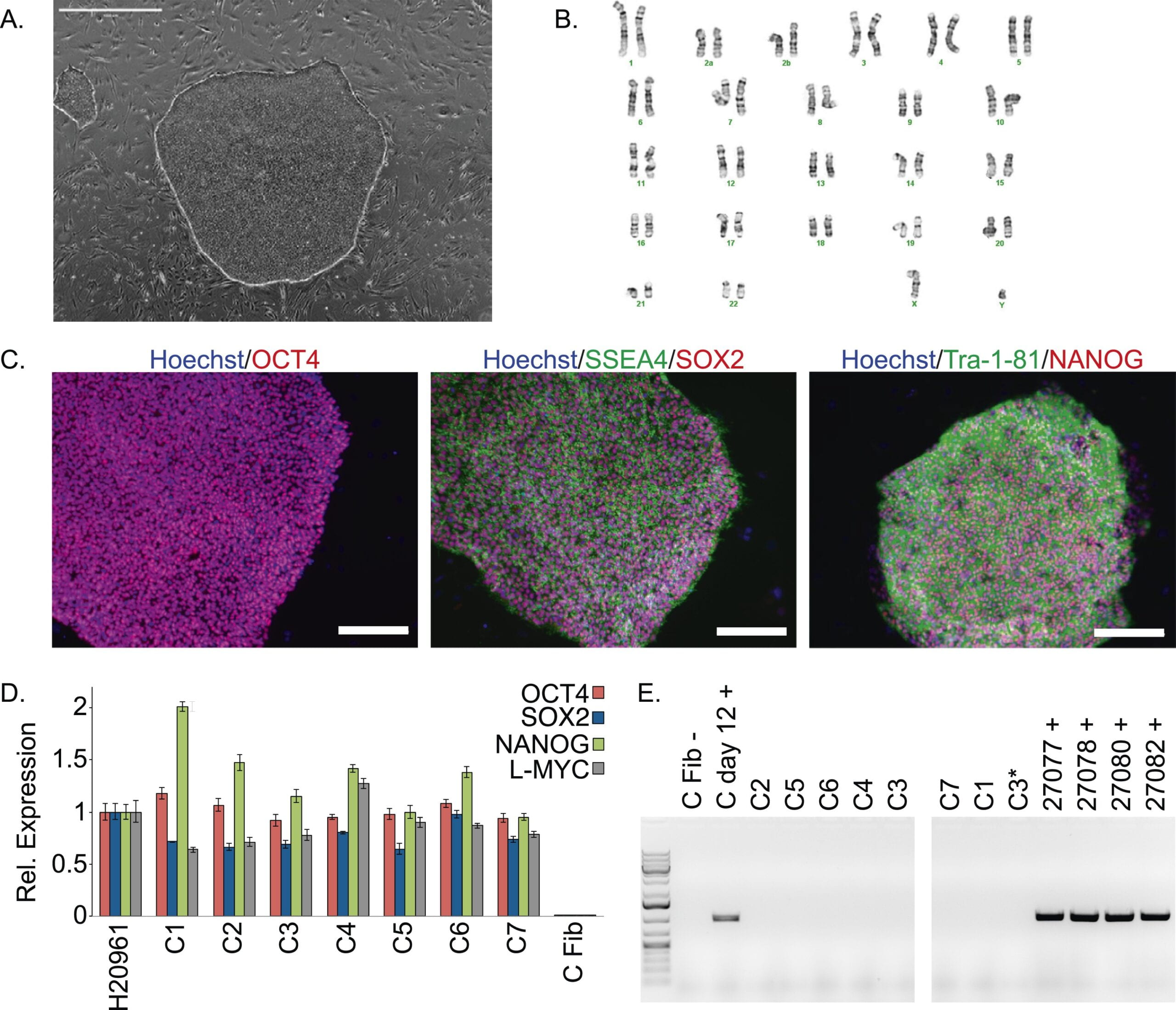
A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics
Gallego Romero I, Pavlovic BJ, Hernando-Herraez I, Zhou X, Ward MC, Banovich NE, Kagan CL, Burnett JE, Huang CH, Mitrano A, Chavarria CI, Friedrich Ben-Nun I, Li Y, Sabatini K, Leonardo TR, Parast M, Marques-Bonet T, Laurent LC, Loring JF, Gilad Y
eLife; 2015
Supplementary data: TXT file
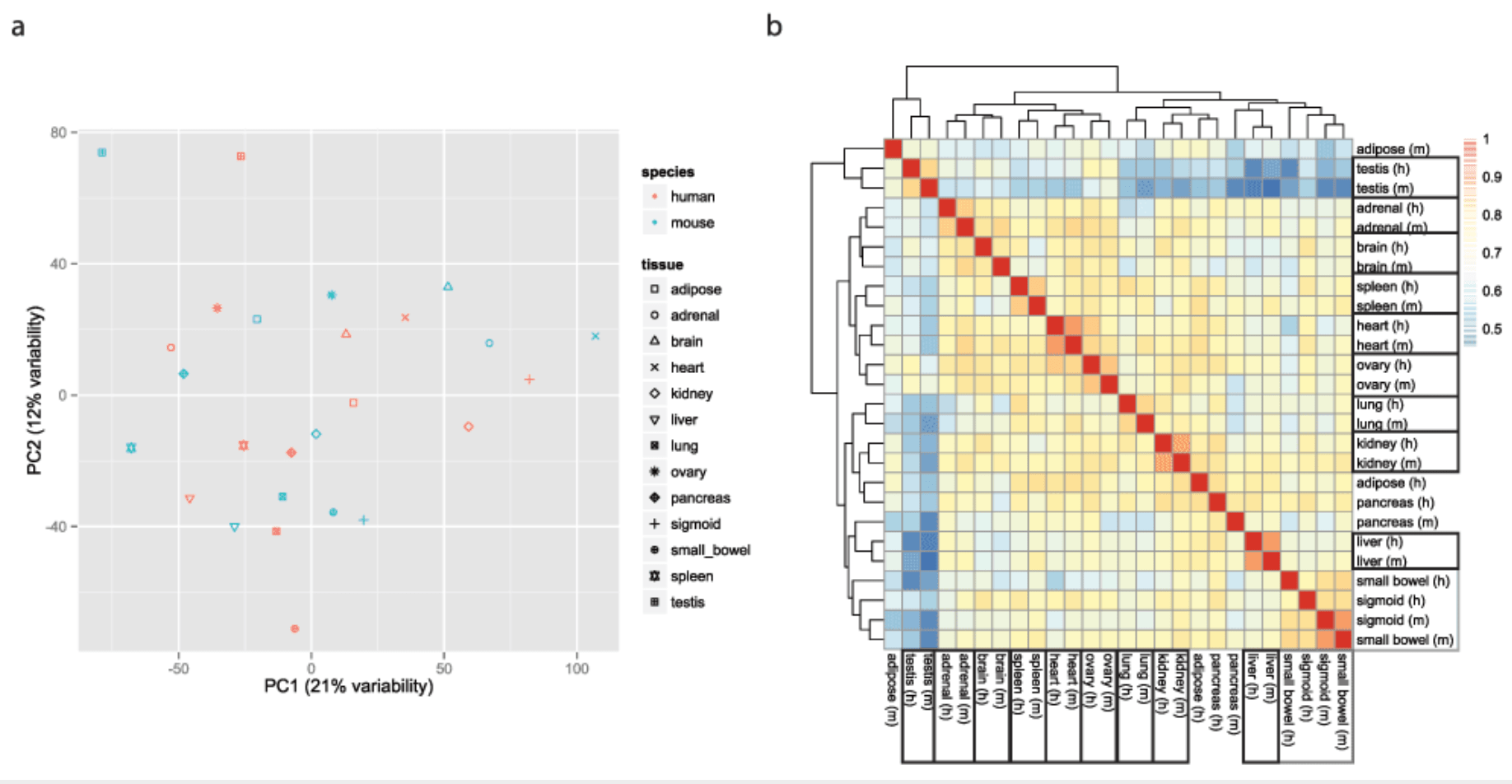
A reanalysis of mouse ENCODE comparative gene expression data
Mizrahi-Man O and Gilad Y
F1000 Research; 2015
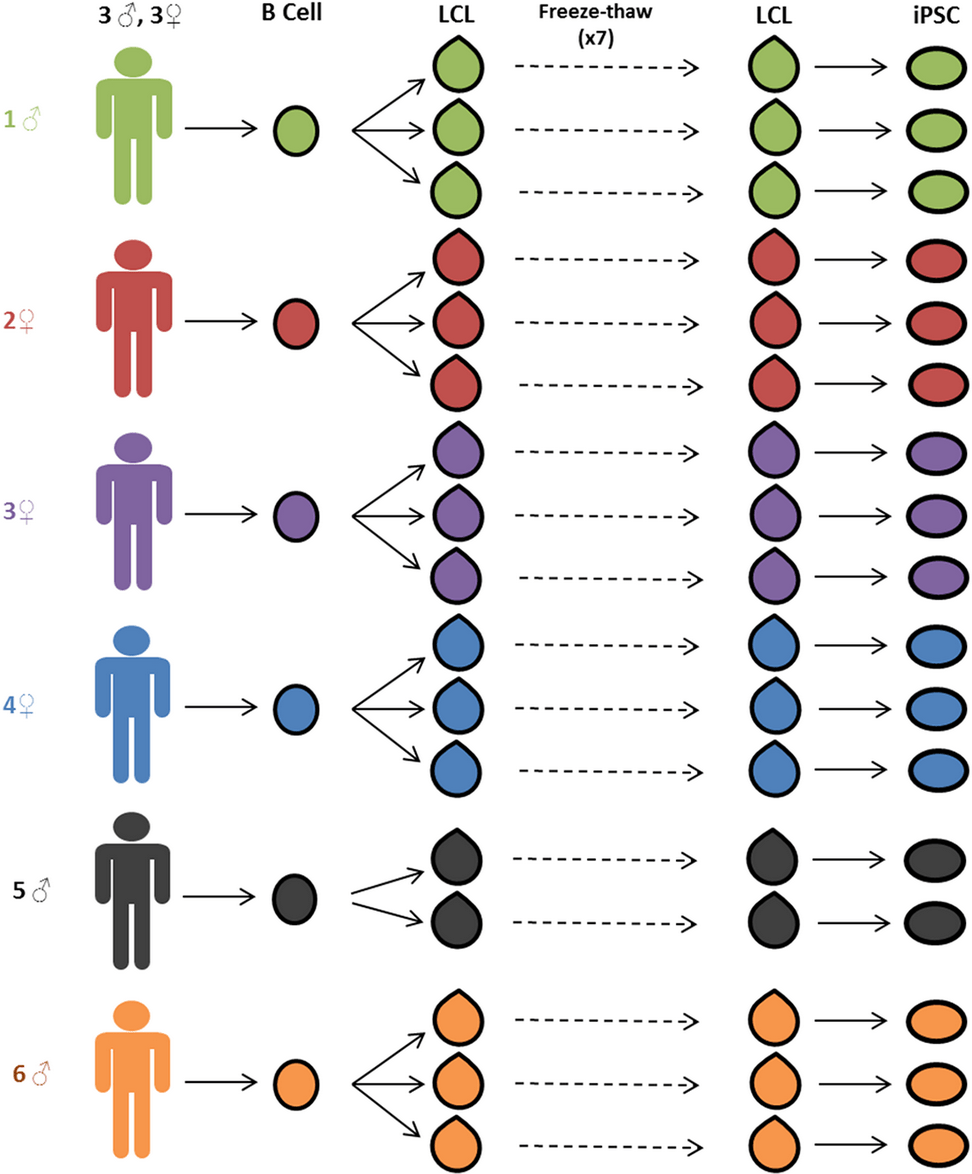
Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
Thomas SM, Kagan C, Pavlovic BJ, Burnett J, Patterson K, Pritchard JK, and Gilad Y
PLoS Genetics; 2015
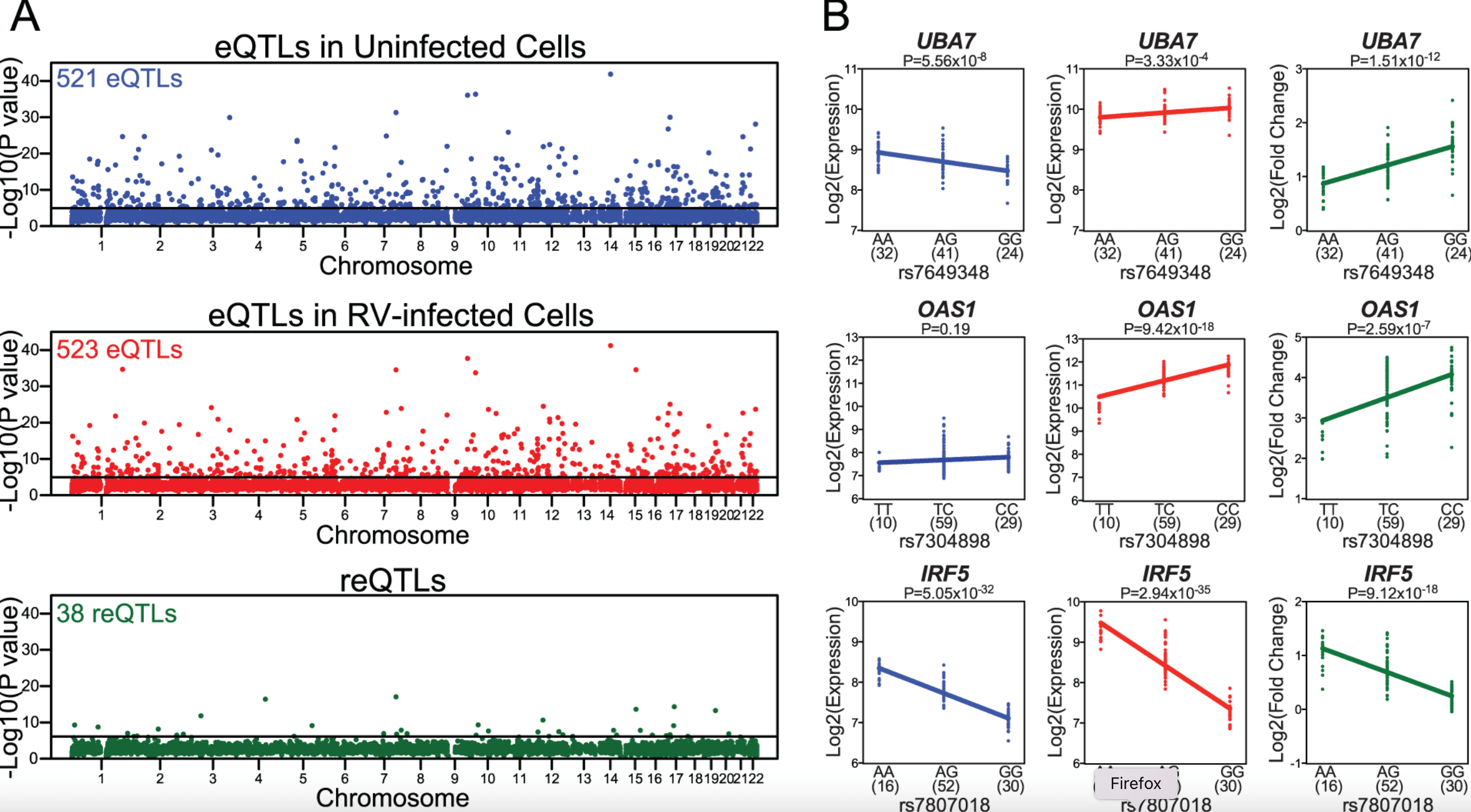
Host Genetic Variation Influences Gene Expression Response to Rhinovirus Infection
Çalışkan M, Baker SW, Gilad Y, and Ober C.
PLoS Genetics; 2015
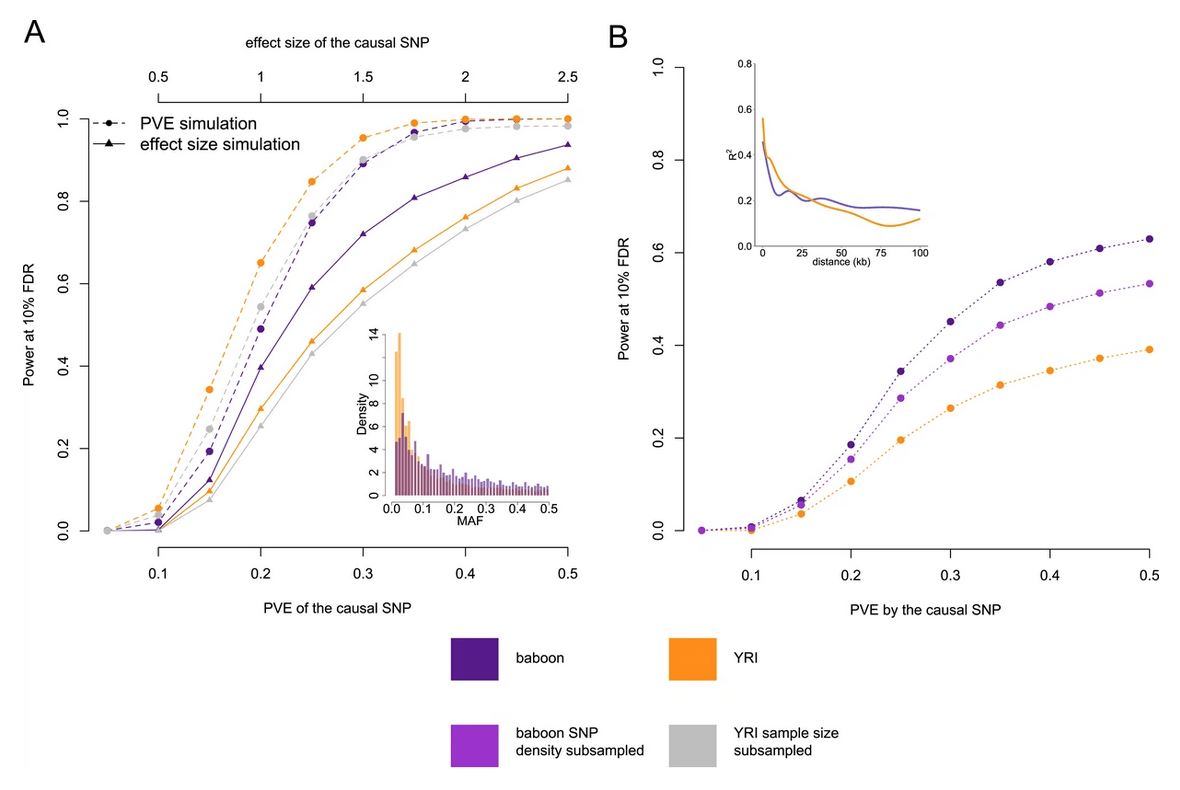
The genetic architecture of gene expression levels in wild baboons
Tung J, Zhou X, Alberts SC, Stephens M, and Gilad Y
eLife; 2015
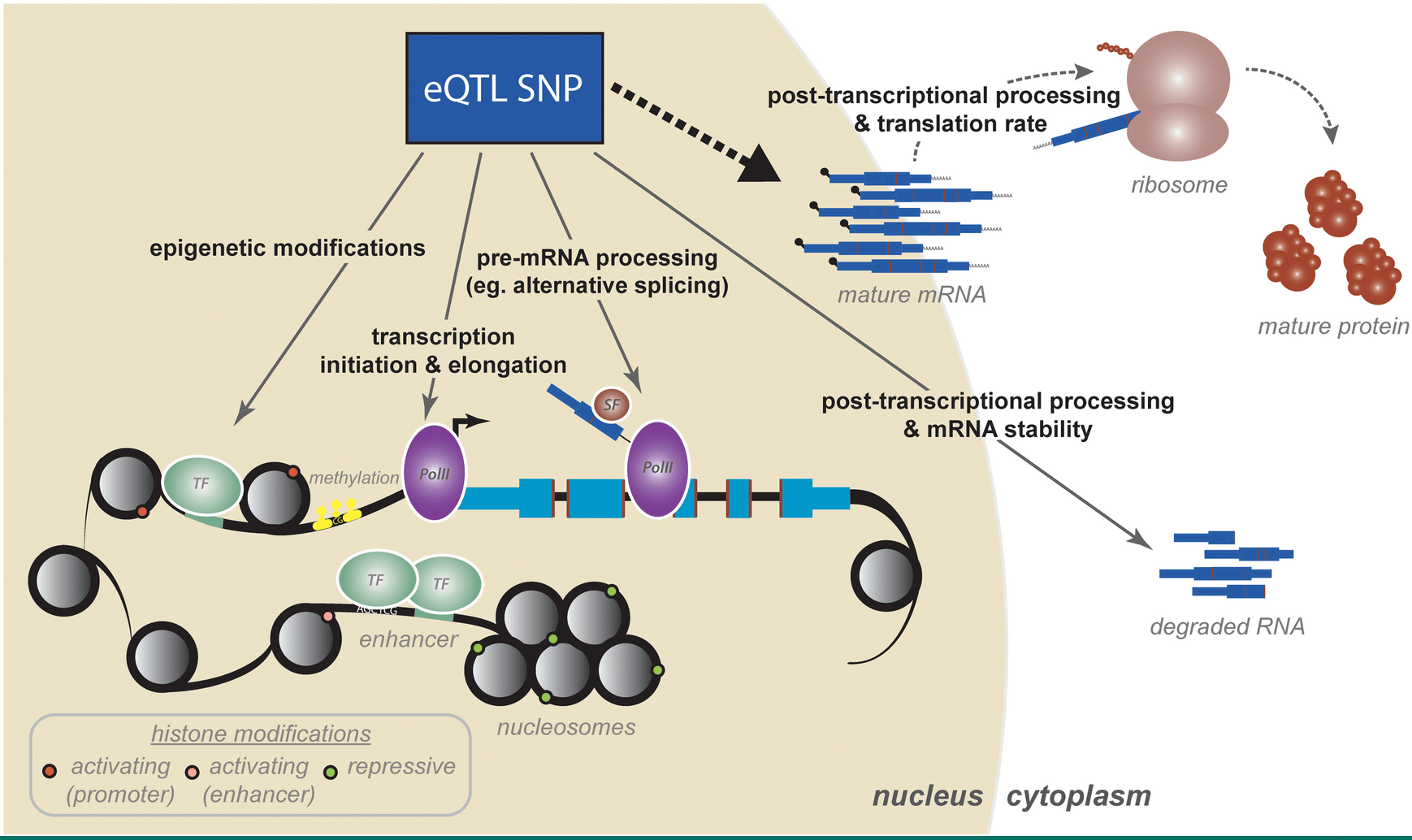
The Genetic and Mechanistic Basis for Variation in Gene Regulation
Pai AA, Pritchard JK, and Gilad Y
PLoS Genetics; 2015
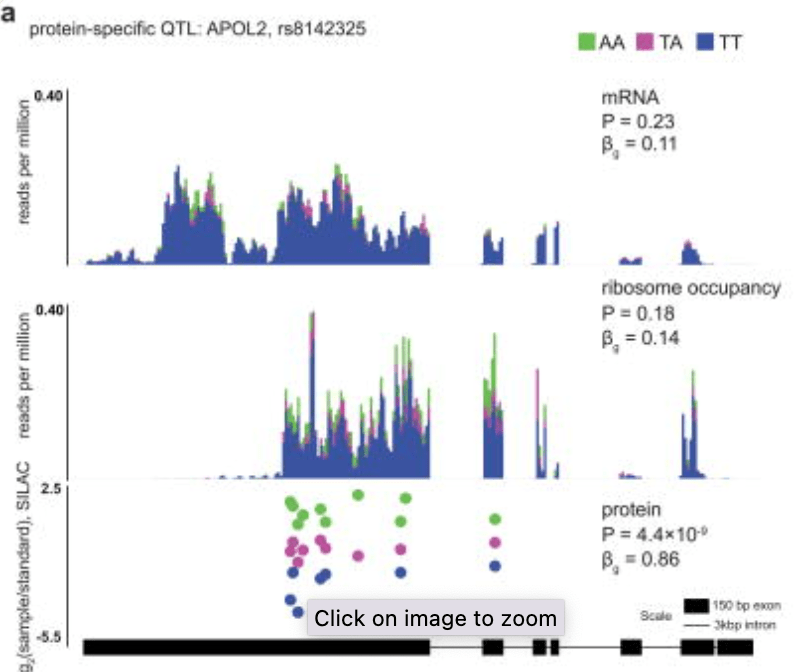
Impact of regulatory variation from RNA to protein
Battle A, Kahn Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, and Gilad Y
Science; 2015
Supplementary data: ZIP file
Genotype data and RNA sequencing data from the 72 YRI LCL lines used in this study are available on GEO (GSE19480 ) and GEUVADIS (E-GEUV-3; ArrayExpress), as is the ribosome profiling data generated in this study (GSE61742). The YRI LCLs have also been used to measure a variety of genomic phenotypes, including H3K27ac (GSE58852), DNA methylation (GSE57483), chromatin accessibility (DNAse-seq; GSE31388), transcriptional elongation rates and initiation frequencies (4sU-seq; GSE75220), RNA decay (GSE37451), and protein levels (PXD001406).
2014
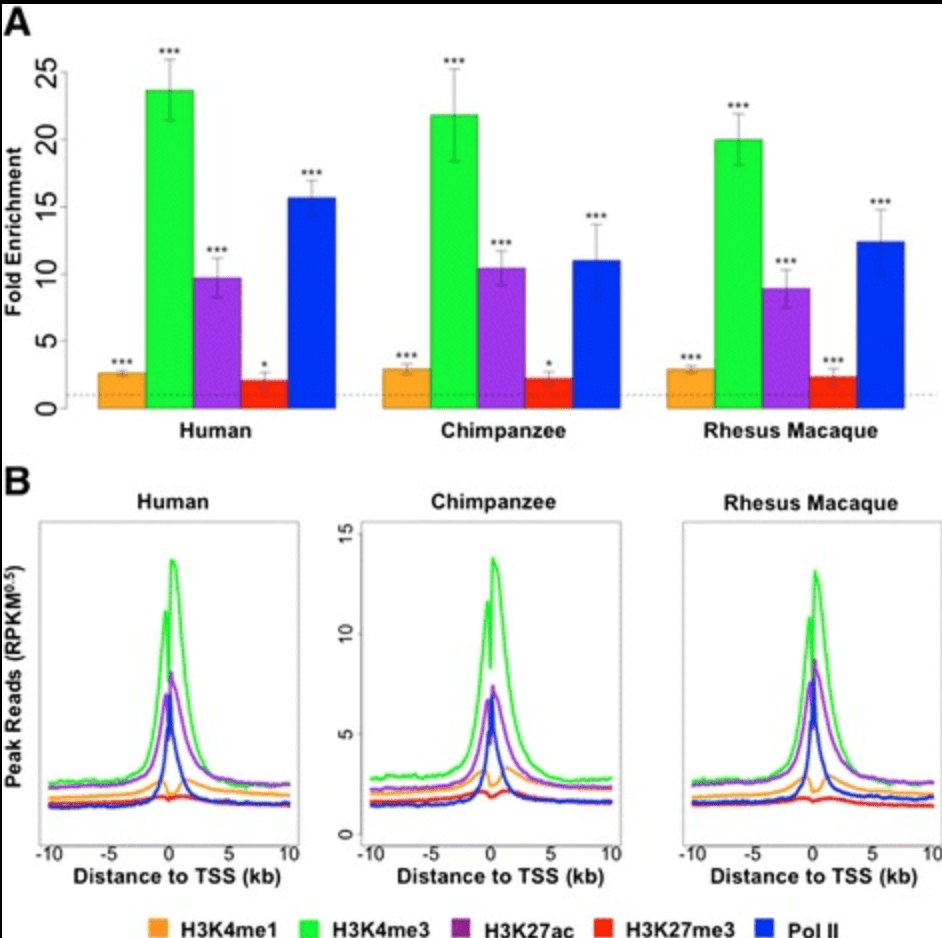
Epigenetic Modifications are Associated with Inter-species Gene Expression Variation in Primates
Zhou X, Cain CE, Myrthil M, Lewellen N, Michelini K, Davenport ER, Stephens M, Pritchard JK, and Gilad Y
Genome Biology; 2014
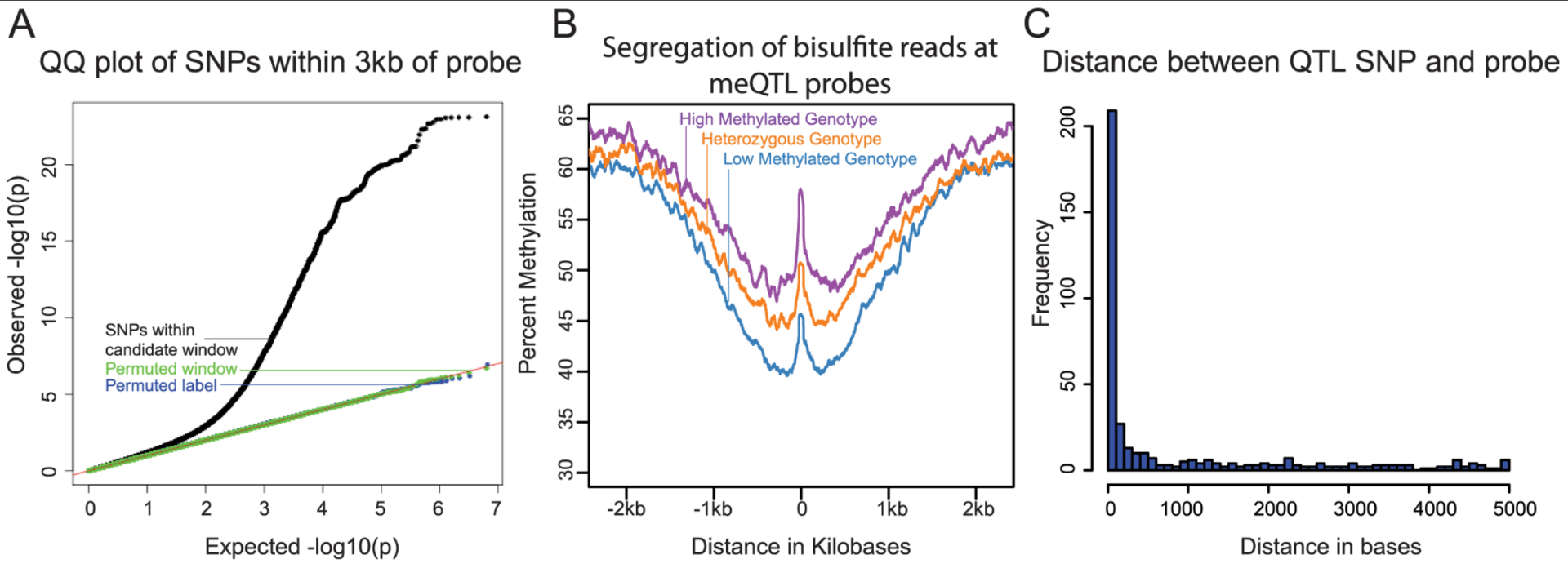
Methylation QTLs Are Associated with Coordinated Changes in Transcription Factor Binding, Histone Modifications, and Gene Expression Levels
Banovich NE, Lan X, McVicker G, van de Geijn B, Degner JF, Blischak JD, Roux J, Pritchard JK, and Gilad Y
PLoS Genetics; 2014
Supplementary data: TXT file
Comparative studies of gene regulatory mechanisms
Pai AA and Gilad Y
The Effect of Freeze-Thaw Cycles on Gene Expression Levels in Lymphoblastoid Cell Lines
Calışkan M, Pritchard JK, Ober C, and Gilad Y
PLoS One; 2014
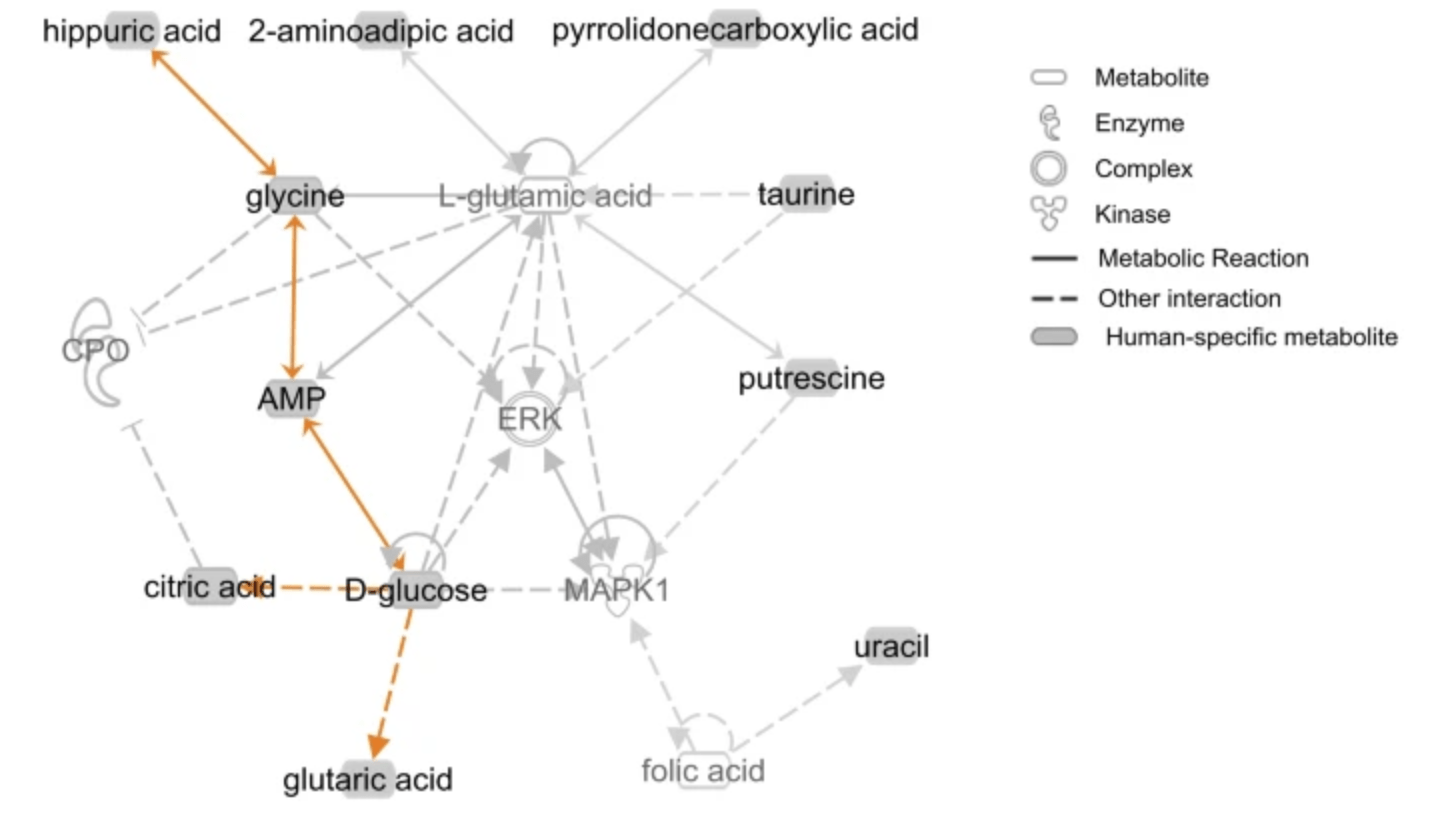
Comparative metabolomics in primates reveals the effects of diet and gene regulatory variation on metabolic divergence
Blekhman R, Perry GH, Shahbaz S, Fiehn O, Clark AC, and Gilad Y
Scientific Reports; 2014

RNA-seq: Impact of RNA degradation on transcript quantification
Gallego Romero I, Pai AA, Tung J, and Gilad Y
BMC Biology; 2014
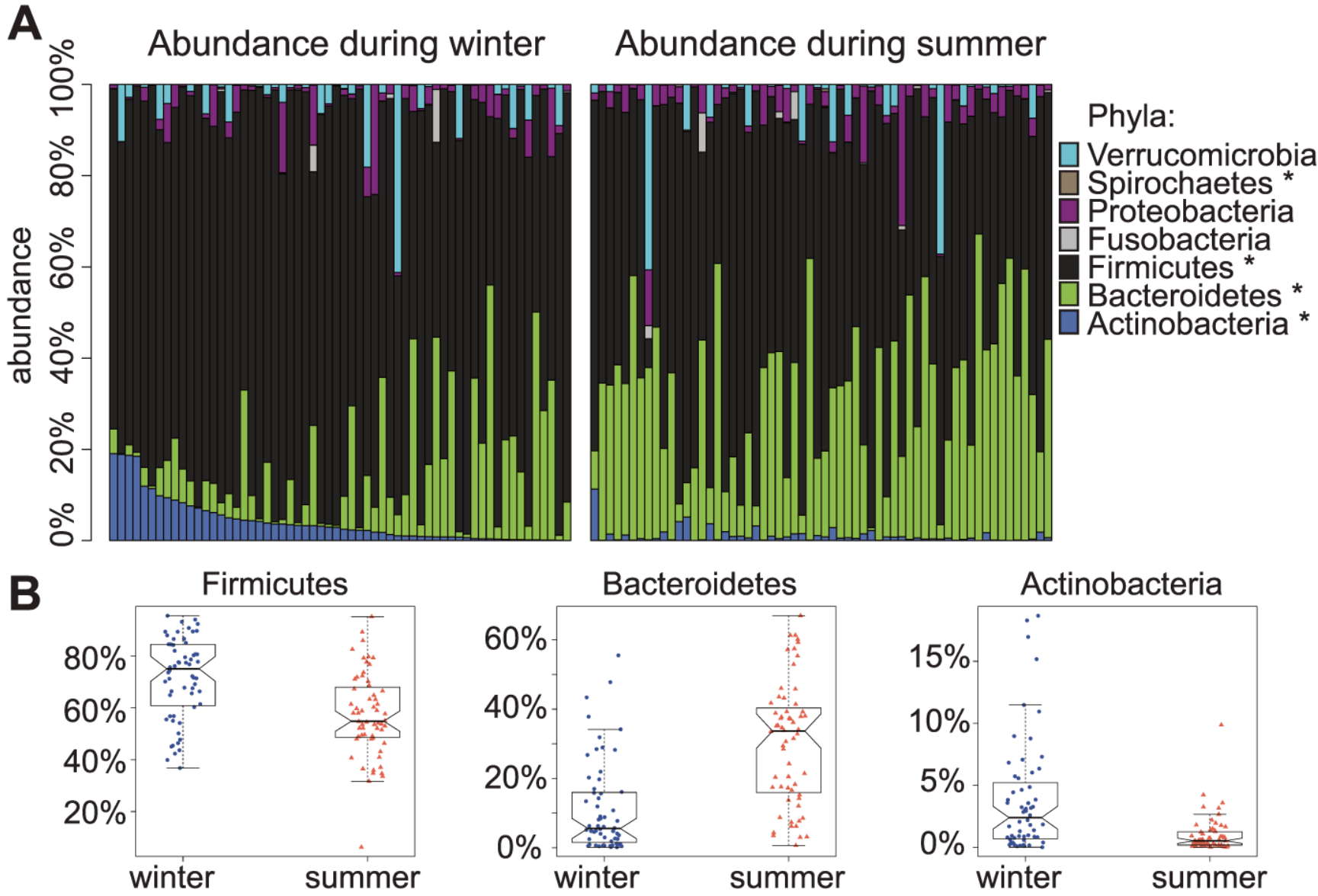
Seasonal variation in human gut microbiome composition
Davenport ER, Mizrahi-Man O, Michelini K, Barreiro LB, Ober C, and Gilad Y
PLoS One; 2014
The functional consequences of variation in transcription factor binding
Cusanovich DA, Pavlovic B, Pritchard JK, and Gilad Y
PLoS Genetics; 2014
Does a unique olfactory genome imply a unique olfactory world?
Frumin I, Sobel N, and Gilad Y
Nature Neuroscience; 2014
2013
Co-binding by YY1 identifies the transcriptionally active, highly conserved set of CTCF-bound regions in primate genomes
Schwalie PC, Ward MC, Cain CE, Faure AJ, Gilad Y, Odom DT, and Flicek P
Genome Biology; 2013
Identification of genetic variants that affect histone modifications in human cells
McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, Lewellen N, Myrthil M, Gilad Y, and Pritchard JK
Science; 2013
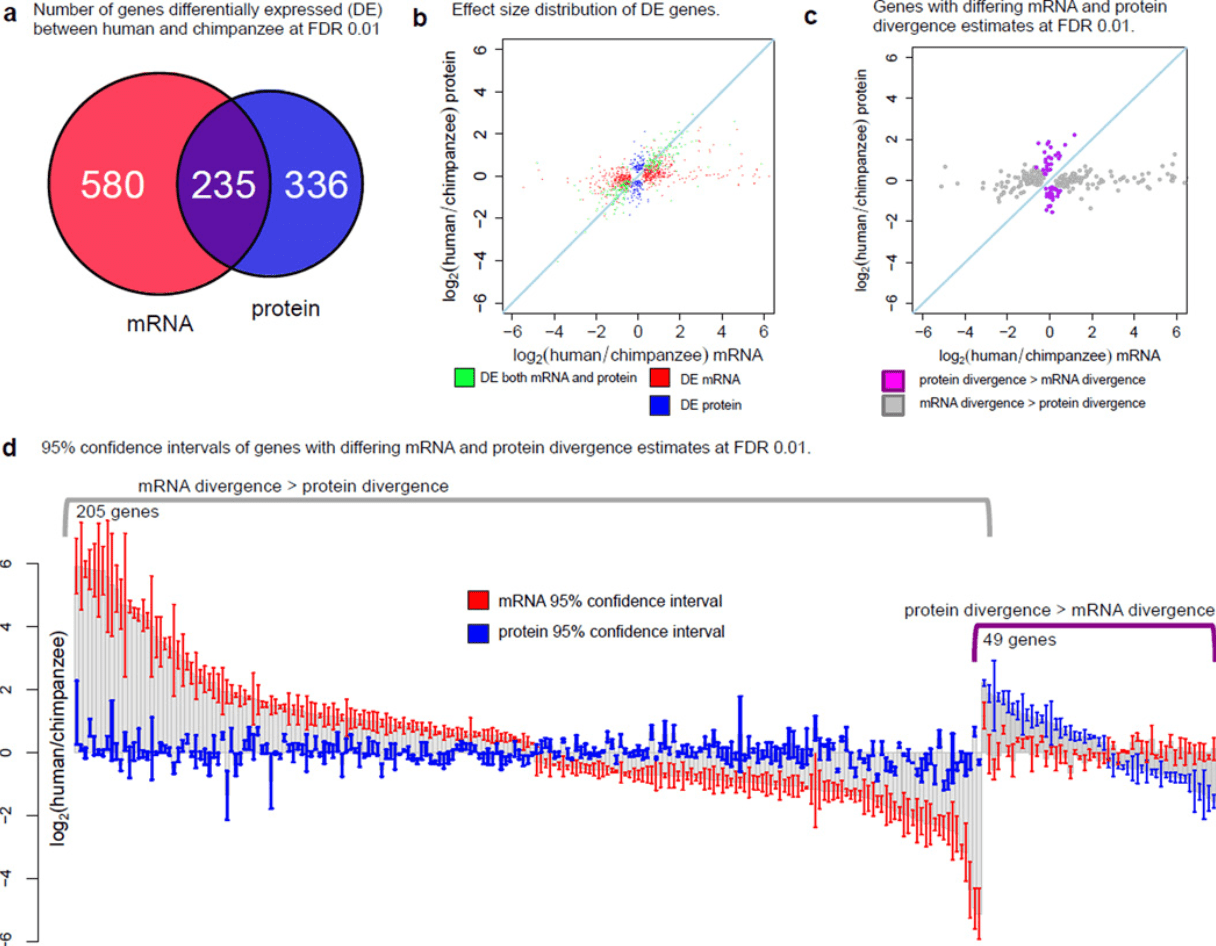
Primate transcript and protein expression levels evolve under compensatory selection pressures
Khan Z, Ford MJ, Cusanovich DA, Mitrano A, Pritchard JK, and Gilad Y
Science; 2013
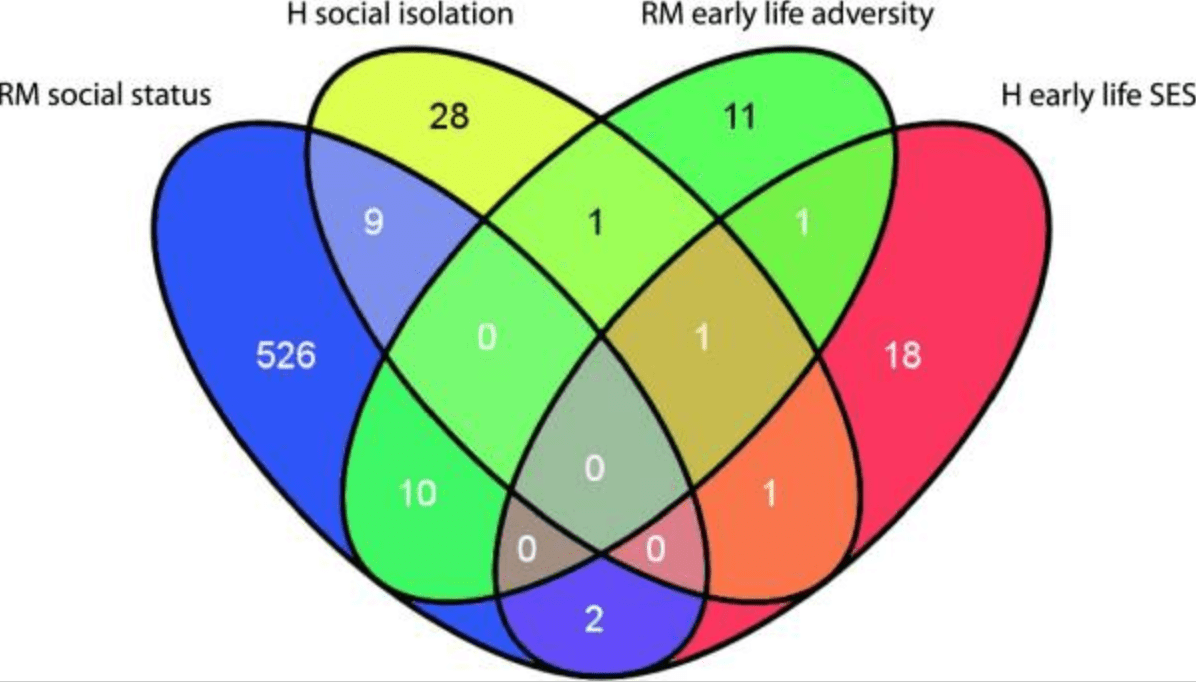
Social environmental effects on gene regulation
Tung J and Gilad Y
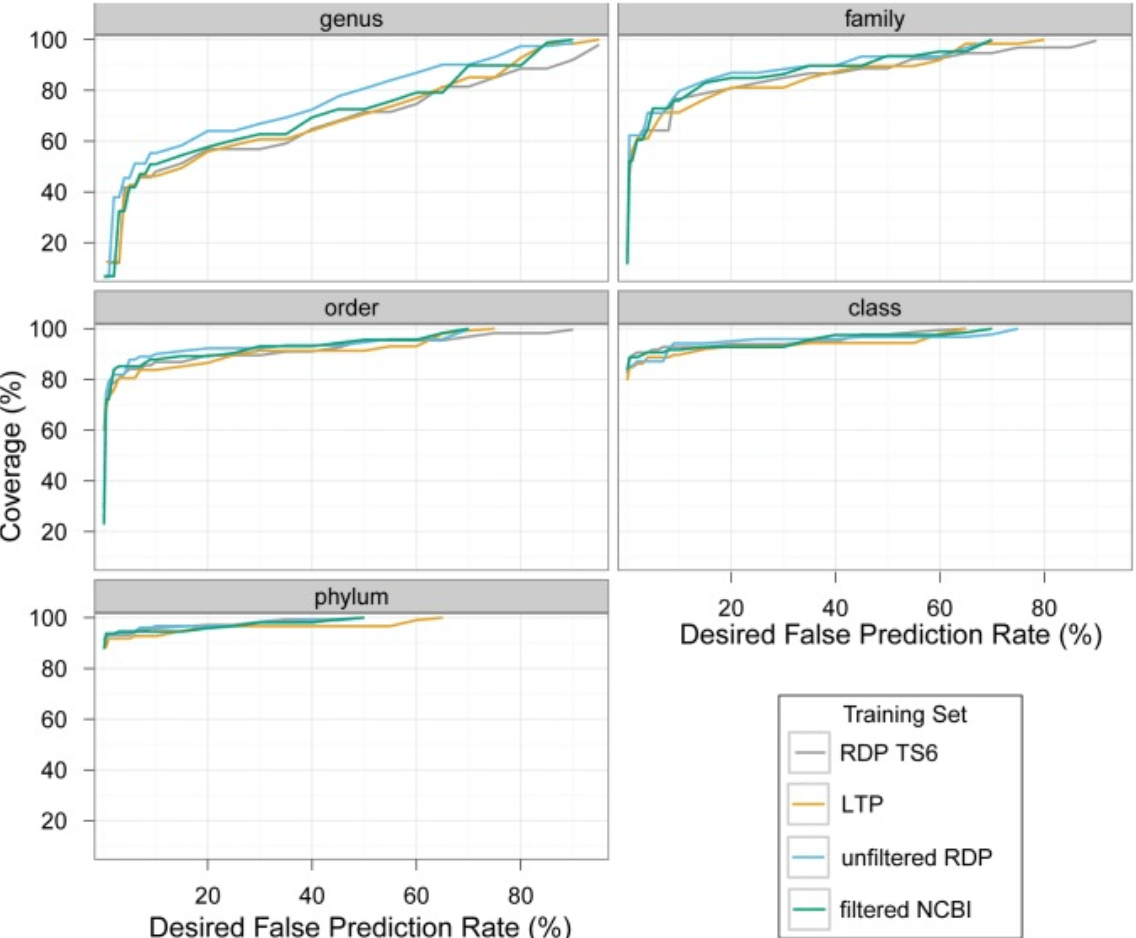
Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: Evaluation of effective study designs
Mizrahi-Man O, Davenport ER, and Gilad Y
PLoS One; 2013
General olfactory sensitivity database (GOSdb): Candidate genes and their genomic variations
Keydar I, Ben-Asher E, Feldmesser E, Nativ N, Oshimoto A, Restrepo D, Matsunami H, Chien MS, Pinto JM, Gilad Y, Olender T, and Lancet D
Human Mutation; 2013
2012
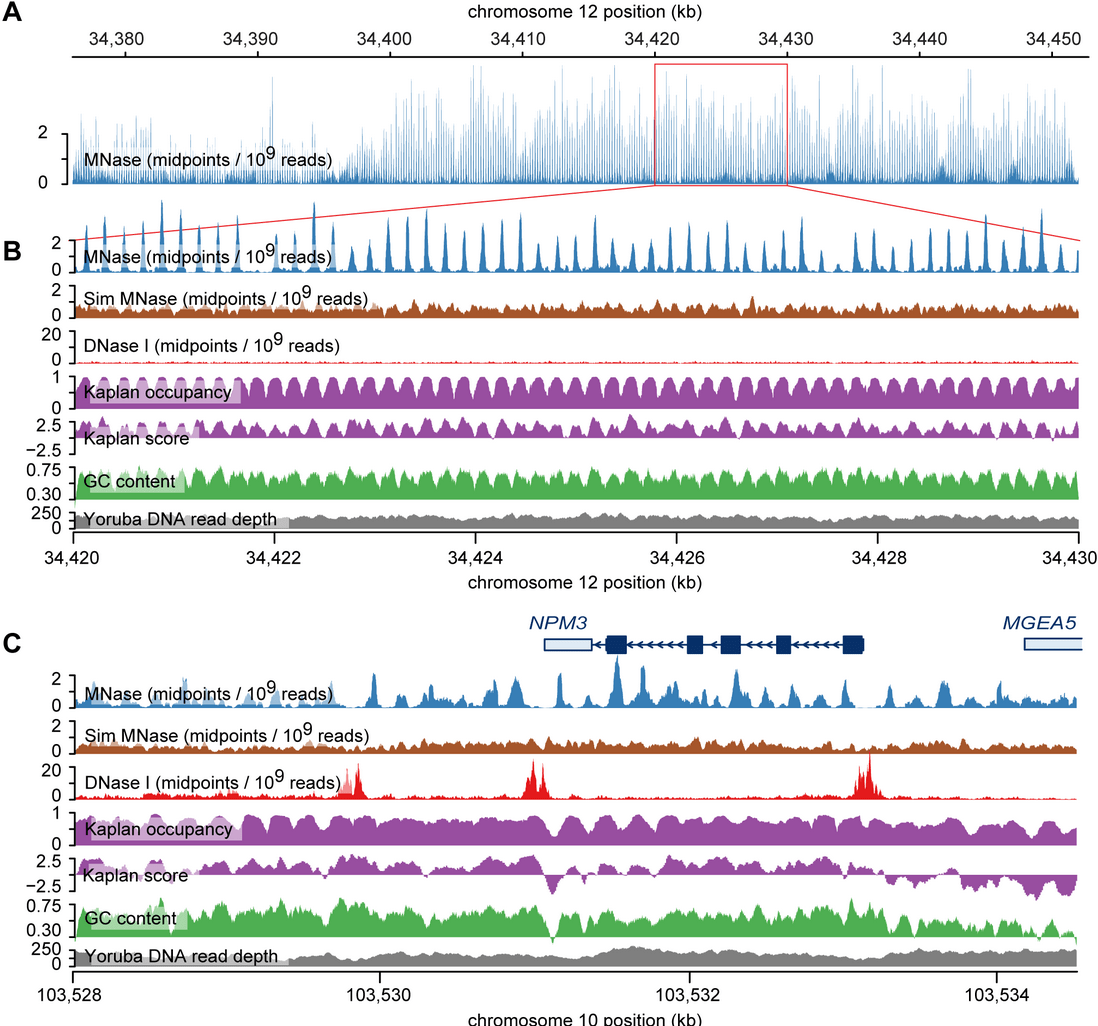
Controls of nucleosome positioning in the human genome
Gaffney DJ, McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, Widom J, Gilad Y, and Pritchard JK
PLoS Genetics; 2012
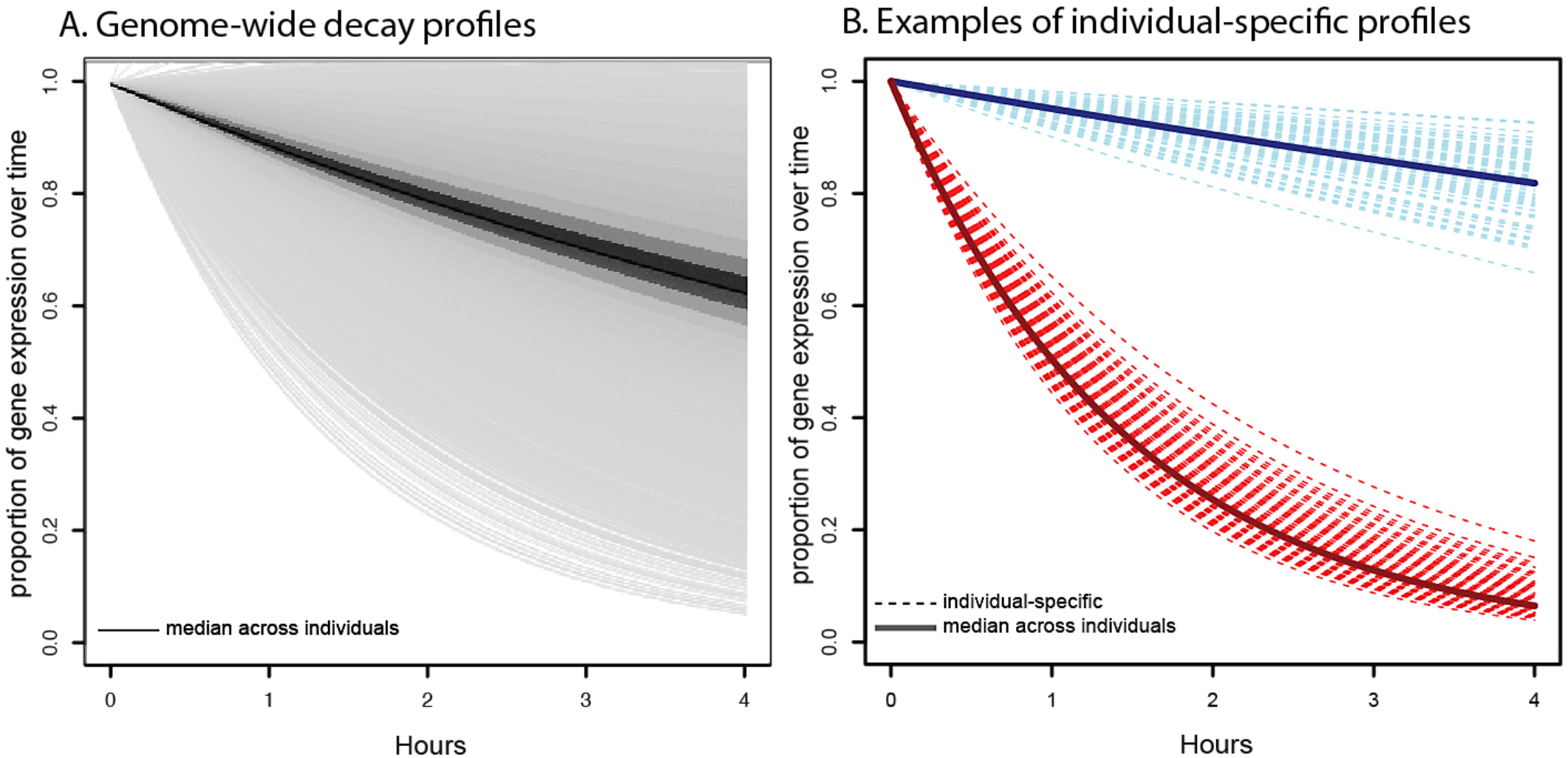
The contribution of RNA decay quantitative trait loci to inter-individual variation in steady-state gene expression levels
Pai AA, Cain CC, Mizrahi-Man O, De Leon S, Lewellen N, Veyrieras JB, Degner JF, Gaffney DJ, Pickrell JK, Stephens M, Pritchard JK, and Gilad Y
PLoS Genetics; 2012
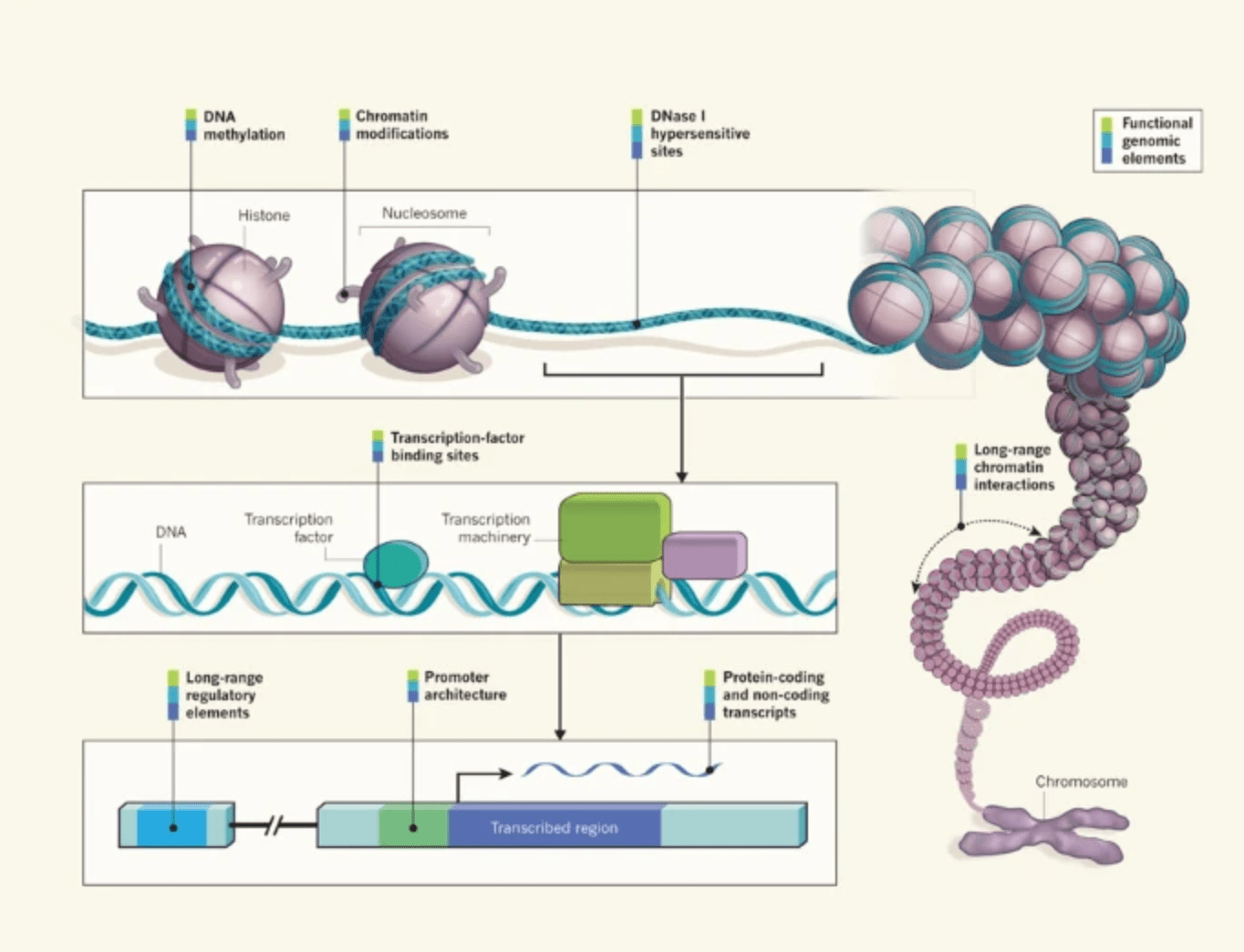
Genomics: ENCODE explained
Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, and Segal E
Nature; 2012
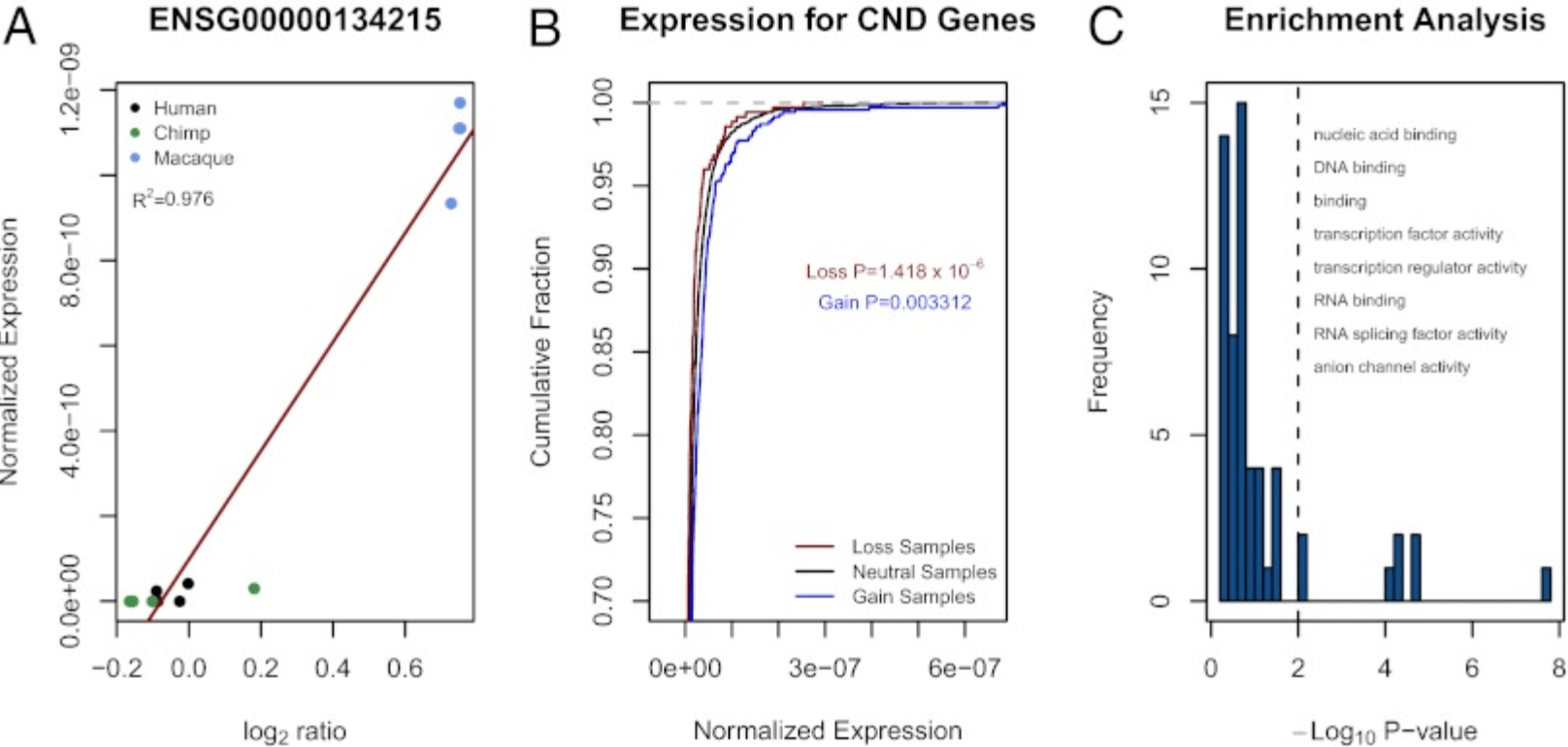
Regulatory element copy number differences shape primate expression profiles
Iskow RC, Gokcumen O, Abyzov A, Malukiewicz J, Zhu Q, Sukumar AT, Pai AA, Mills RE, Habegger L, Cusanovich DA, Rubel MA, Perry GH, Gerstein M, Stone AC, Gilad Y, and Lee C
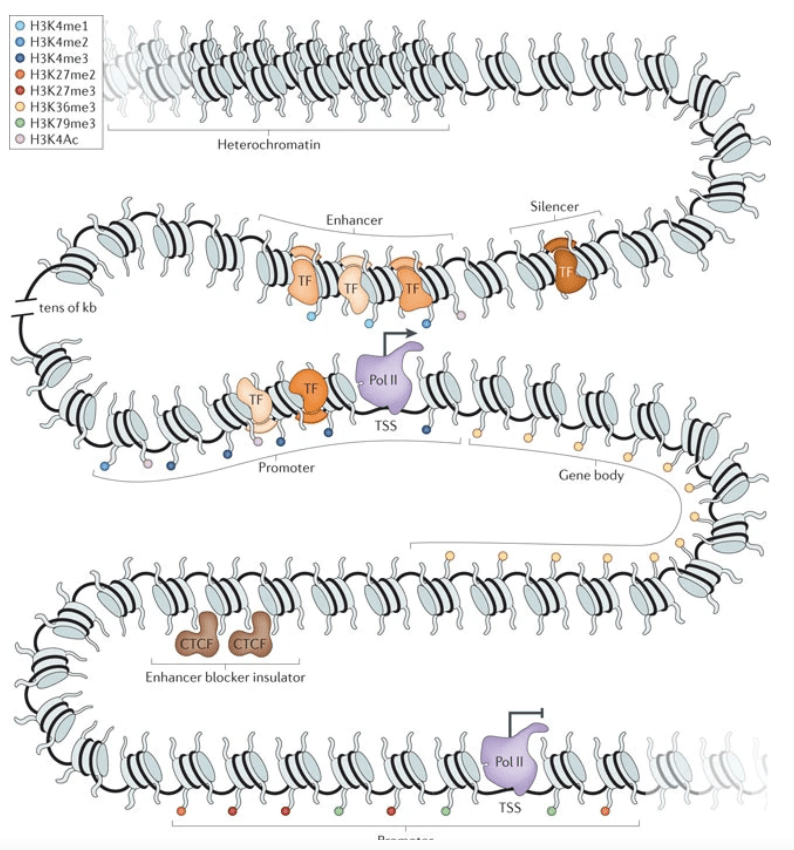
Comparative studies of gene expression and the evolution of gene regulation
Gallego Romero I, Ruvisnky I, and Gilad Y
Nature Reviews Genetics; 2012
Time-dependent transcriptional profiling links gene expression to mitogen-activated protein kinase kinase 4 (MKK4)-mediated suppression of omental metastatic colonization
Bainer RO, Veneris JT, Yamada SD, Montag A, Lingen MW, Gilad Y, and Rinker-Schaeffer CW
The combination of a genome-wide association study of lymphocyte count and analysis of gene expression data reveals novel asthma candidate genes
Cusanovich DA, Billstrand C, Zhou X, Chavarria C, De Leon S, Michelini K, Pai AA, Ober C, and Gilad Y
Human Molecular Genetics; 2012
Social environment is associated with gene regulatory variation in the rhesus macaque immune system
Tung J, Barreiro LB, Johnson ZP, Hansen KD, Michopoulos V, Toufexis D, Michelini K, Wilson ME, and Gilad Y
Proceedings of the National Academy of Sciences; 2012
Supplementary data: DOC file

Comment on “Widespread RNA and DNA sequence differences in the human transcriptome”
Pickrell JK, Gilad Y, and Pritchard JK
Science; 2012
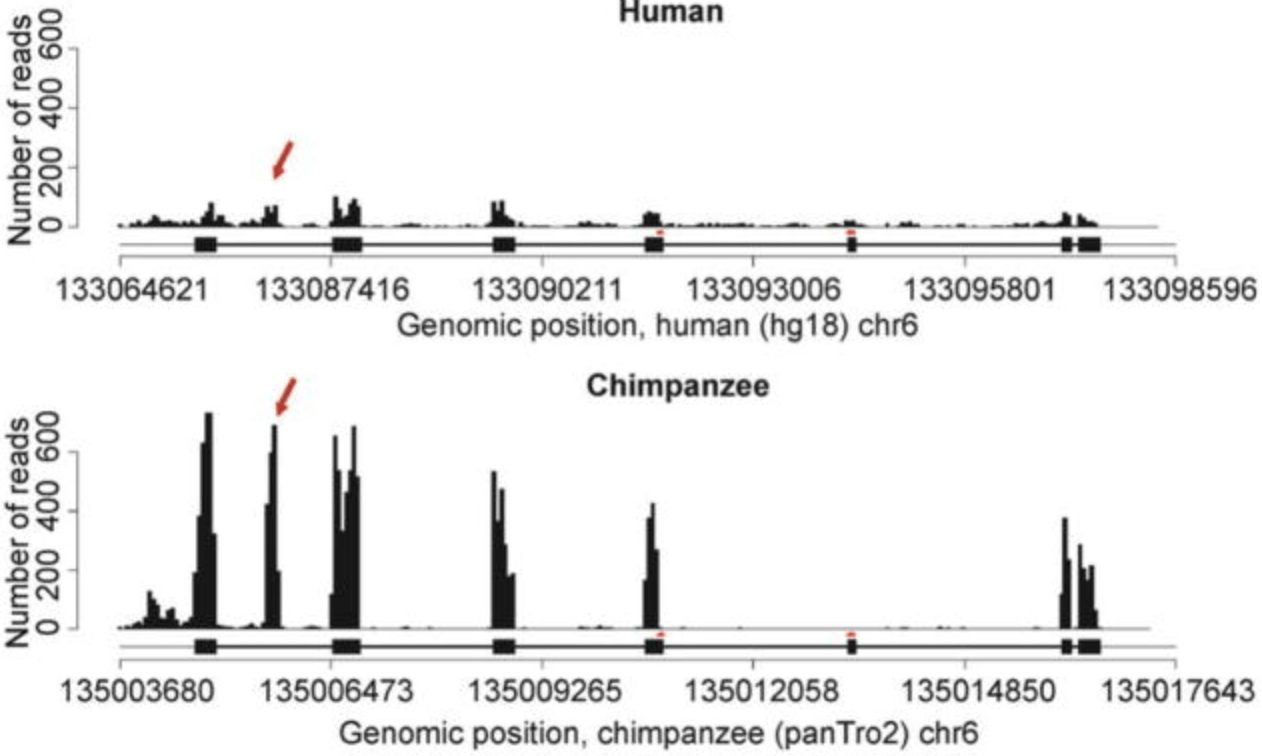
Exon-specific QTLs skew the inferred distribution of expression QTLs detected using gene expression array data
Veyrieras JB, Gaffney DJ, Pickrell JK, Gilad Y, Stephens M, and Pritchard JK
PLoS One; 2012
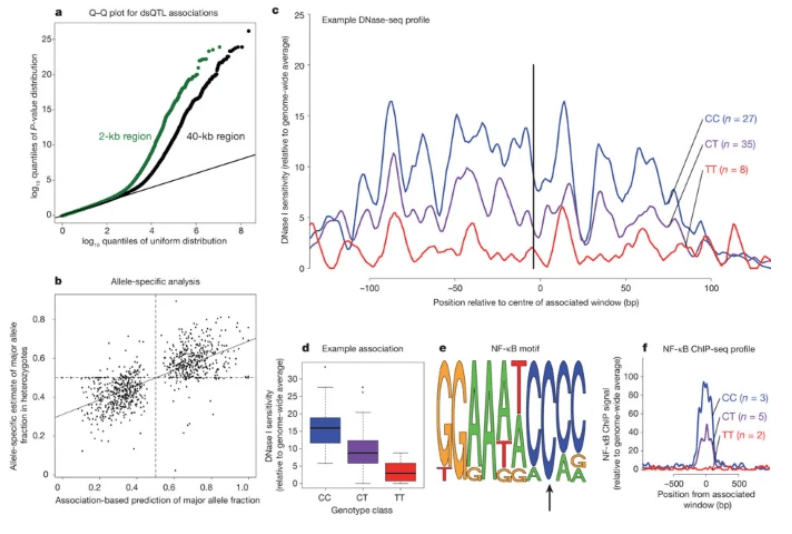
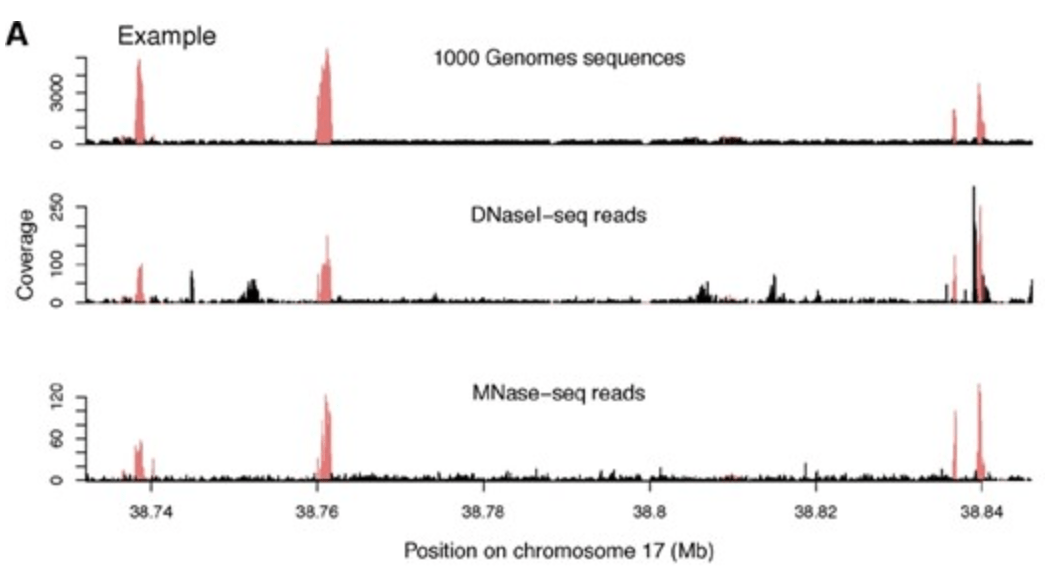
DNase I sensitivity QTLs are a major determinant of human expression variation
Degner JF, Pai AA, Pique-Regi R, Veyrieras JB, Gaffney DJ, Pickrell JK, De Leon S, Michelini K, Lewellen N, Crawford GE, Stephens M, Gilad Y, and Pritchard JK
Nature; 2012
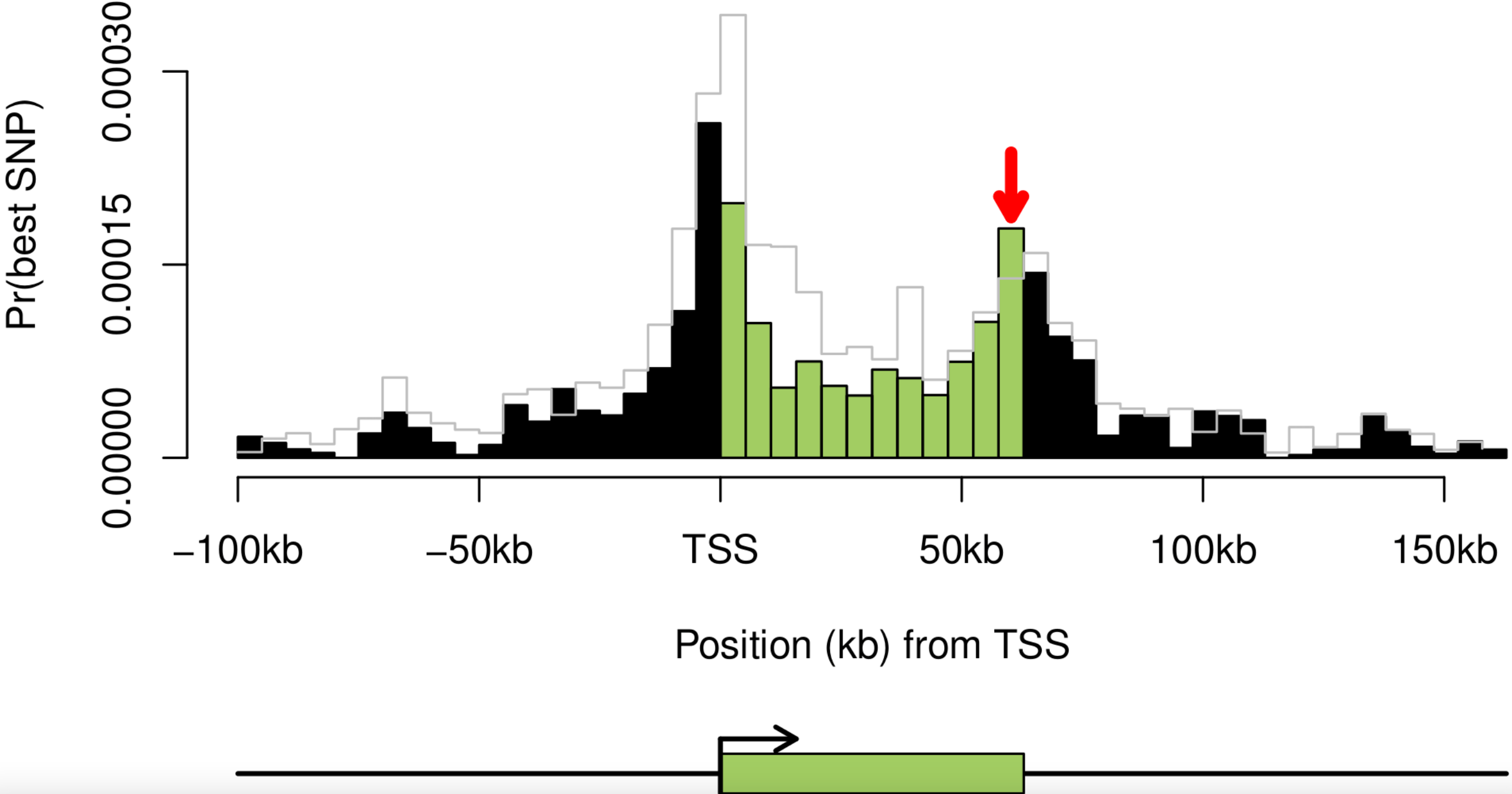
Dissecting the regulatory architecture of gene expression QTLs
Gaffney DJ, Veyrieras JB, Degner JF, Pique-Regi R, Pai AA, Crawford GE, Stephens M, Gilad Y, and Pritchard JK
Genome Biology; 2012
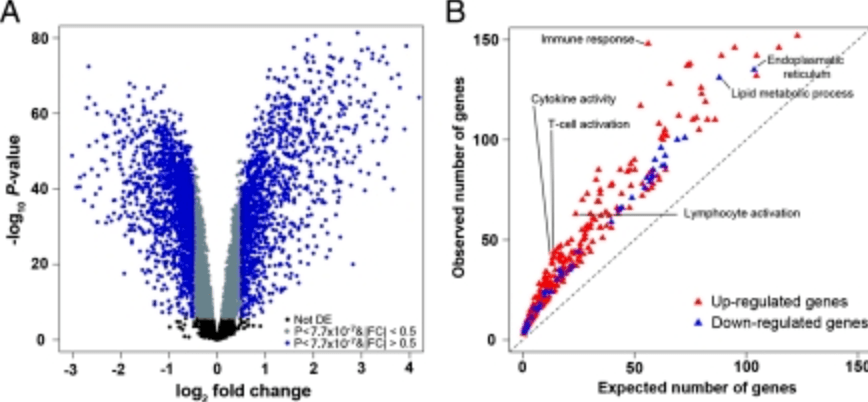
Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection
Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, and Gilad Y
Comparative RNA sequencing reveals substantial genetic variation in endangered primates
Perry GH, Melsted P, Marioni JC, Wang Y, Bainer R, Pickrell JK, Michelini K, Zehr S, Yoder AD, Stephens M, Pritchard JK, and Gilad Y
Genome Research; 2012
Supplemental data: Please contact us if data on Genome Research webpage doesn’t meet your needs.
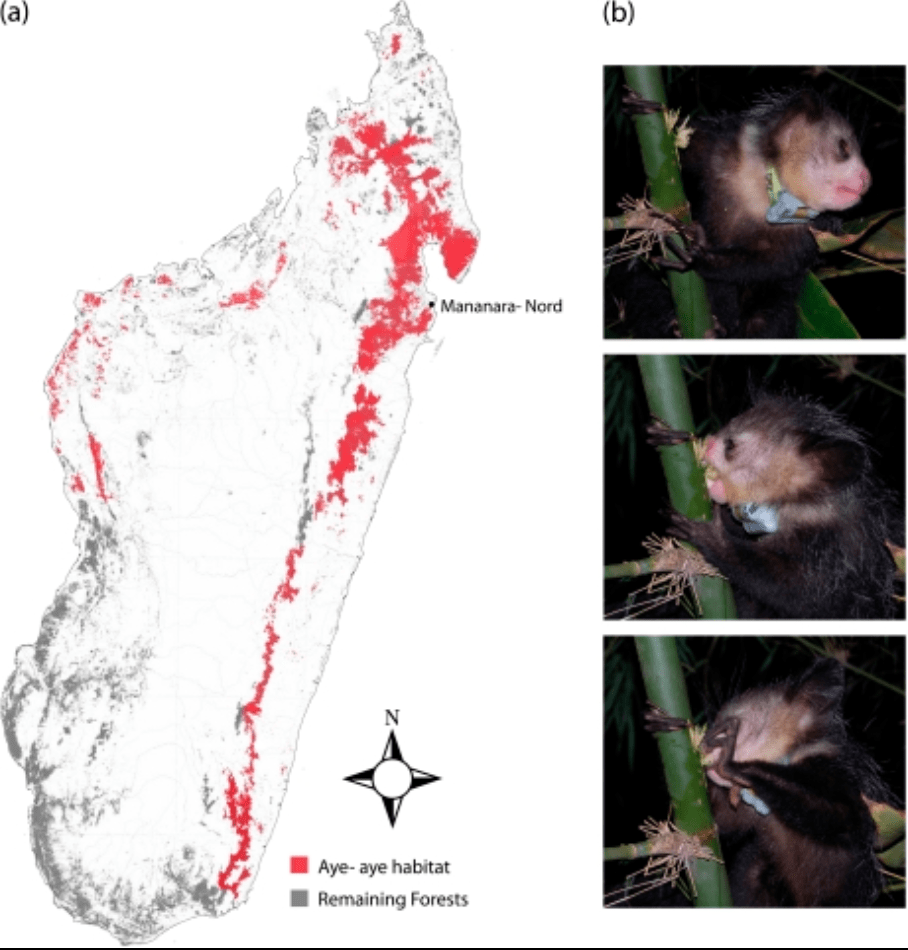
A genome sequence resource for the aye-aye (Daubentonia madagascariensis), a nocturnal lemur from Madagascar
Perry GH, Reeves D, Melsted P, Ratan A, Miller W, Michelini K, Louis EE Jr, Pritchard JK, Mason CE, and Gilad Y
Genome Biology and Evolution; 2012
Supplemental data: ZIP file
2011
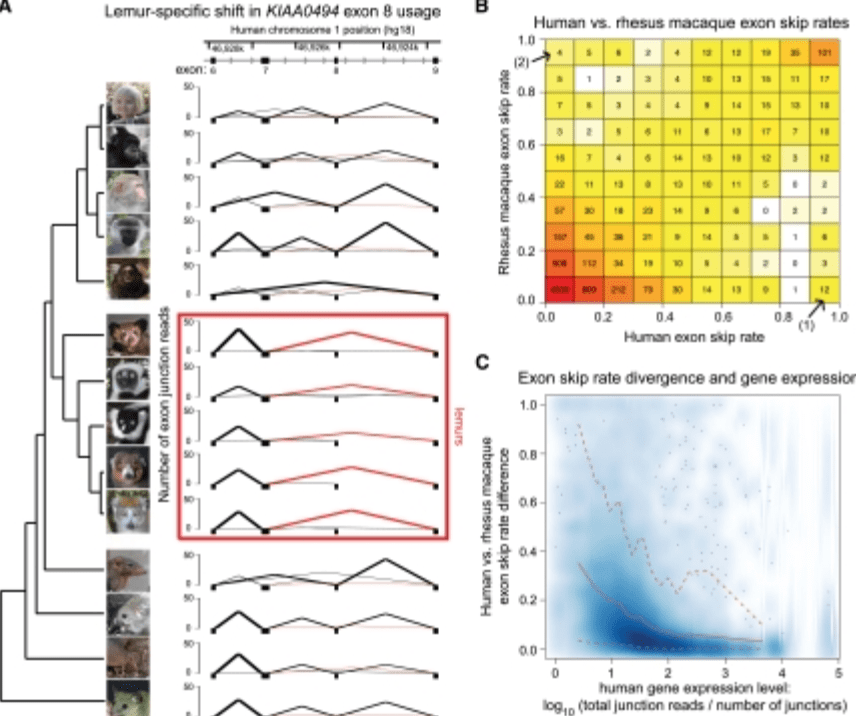
False positive peaks in ChIP-seq and other sequencing-based functional assays caused by unannotated high copy number regions
Pickrell JK, Gaffney DJ, Gilad Y, and Pritchard JK
Bioinformatics; 2011

A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues
Pai AA, Bell JT, Marioni JC, Pritchard JK, and Gilad Y
PLoS Genetics; 2011
Gene expression differences among primates are associated with changes in a histone epigenetic modification
Cain CE, Blekhman R, Marioni JC, and Gilad Y
Genetics; 2011
The effects of EBV transformation on gene expression levels and methylation profiles
Çalışkan M, Cusanovich DA, Ober C, and Gilad Y
Human Molecular Genetics; 2011
Supplemental data: Multiple files types
DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines
Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y and Pritchard JK
Genome Biology; 2011
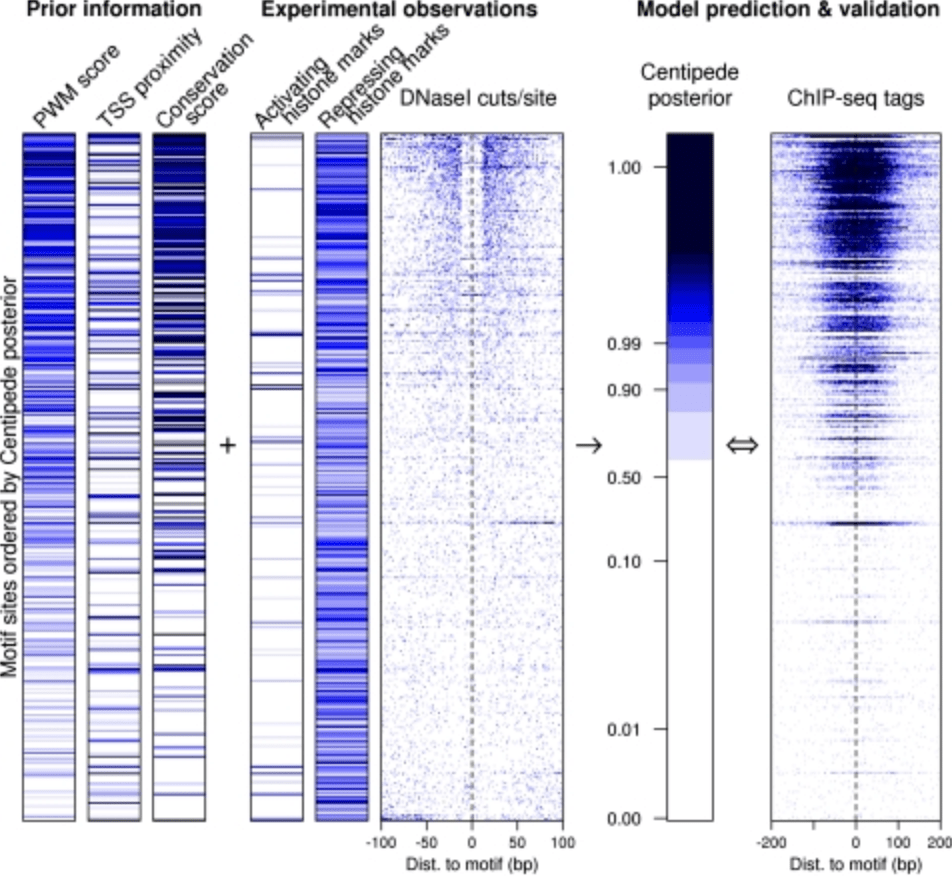
Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data
Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, and Pritchard JK
Genome Research; 2011
2010
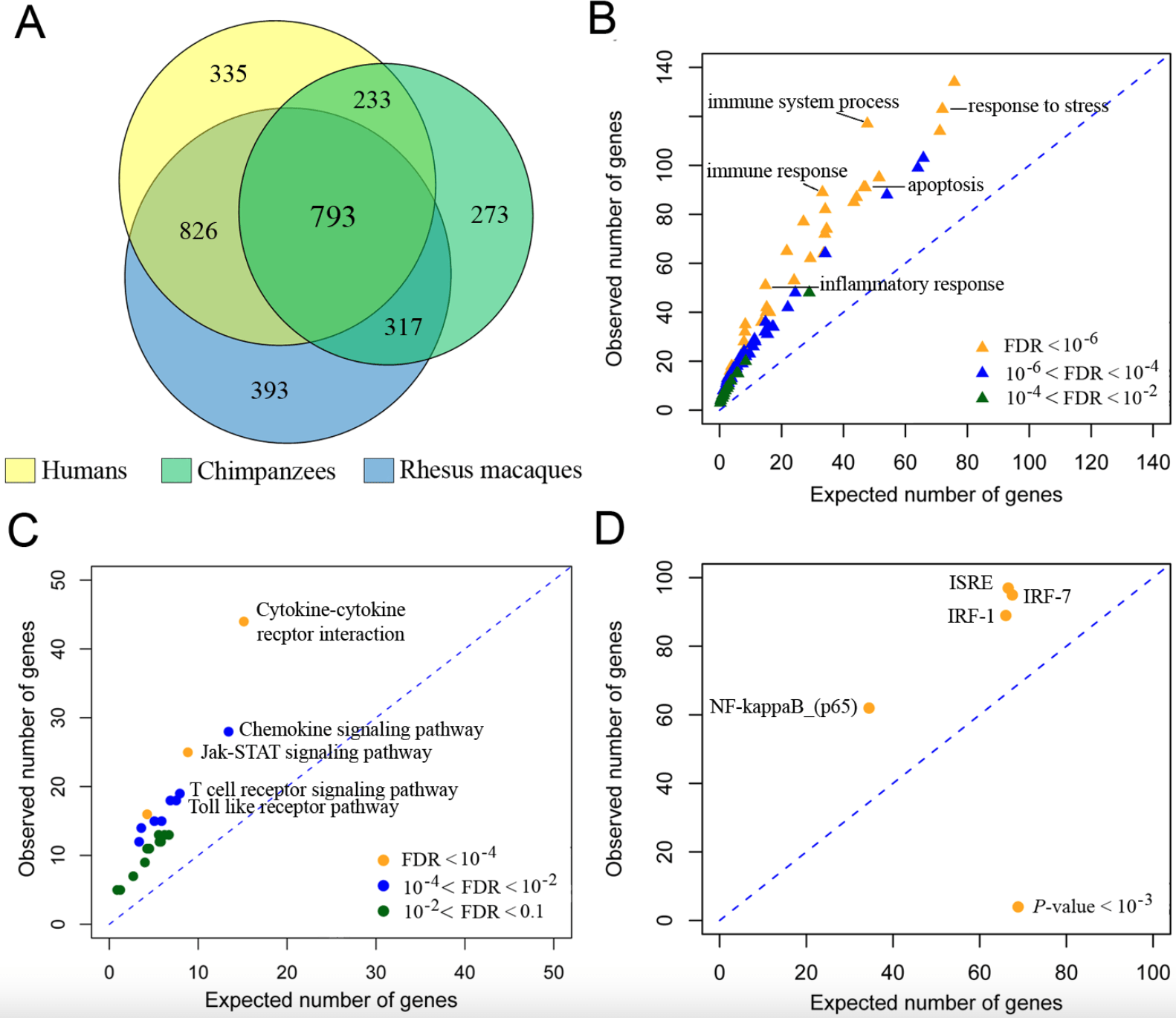
Functional comparison of innate immune signaling pathways in primates
Barreiro LB, Marioni JC, Blekhman R, Stephens M, and Gilad Y
PLoS Genetics; 2010
Noisy splicing drives mRNA isoform diversity in human cells
Pickrell JK, Pai AA, Gilad Y, and Pritchard JK
PLoS Genetics; 2010
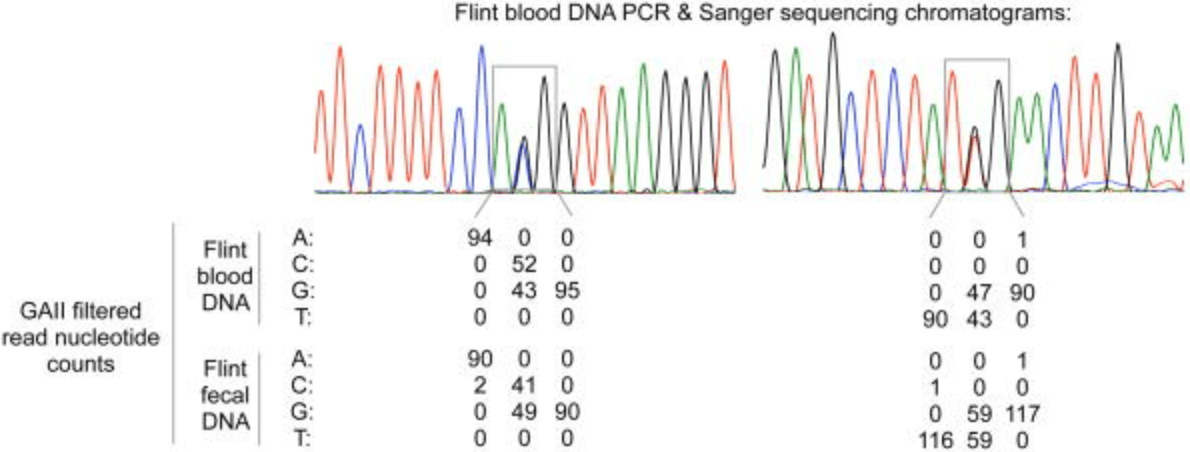
Genomic-scale capture and sequencing of endogenous DNA from feces
Perry GH, Marioni JC, Melsted P, and Gilad Y
Molecular Ecology; 2010
Supplemental data: NIH (study number SRA012374)
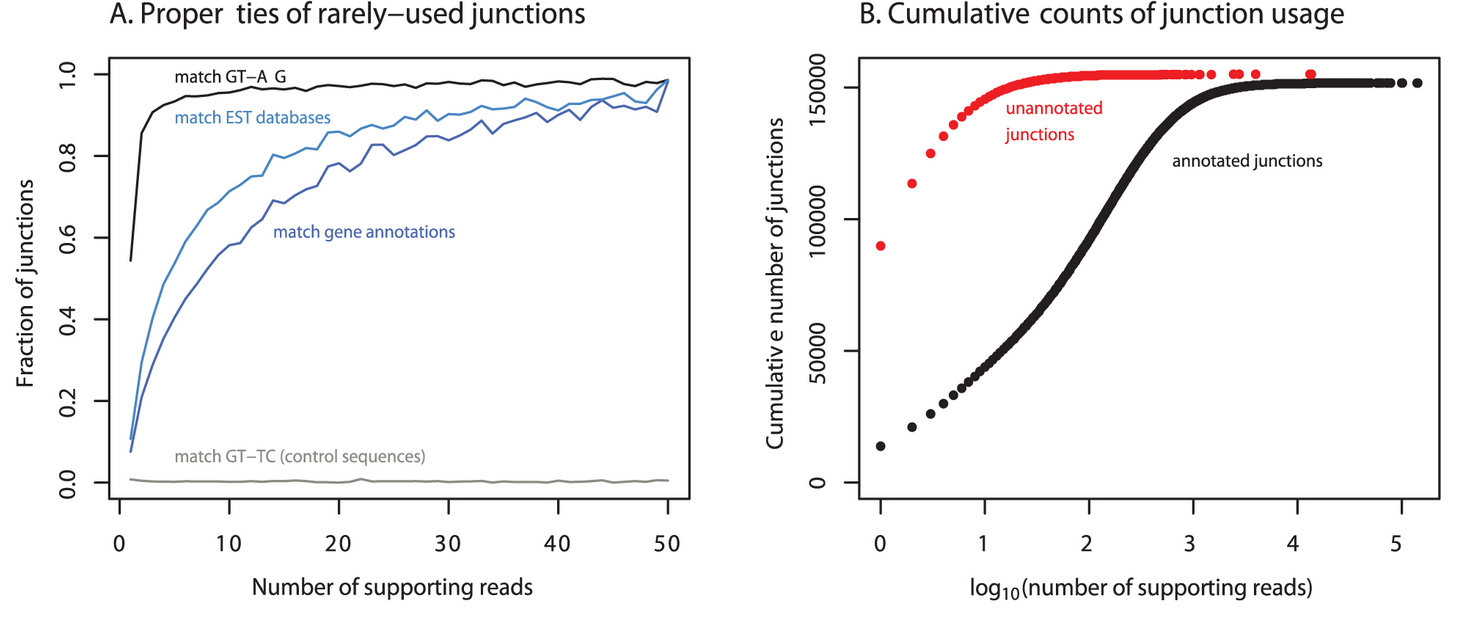
Understanding mechanisms underlying human gene expression variation with RNA sequencing
Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras J-B, Stephens M, Gilad Y, and Pritchard JK
Nature; 2010
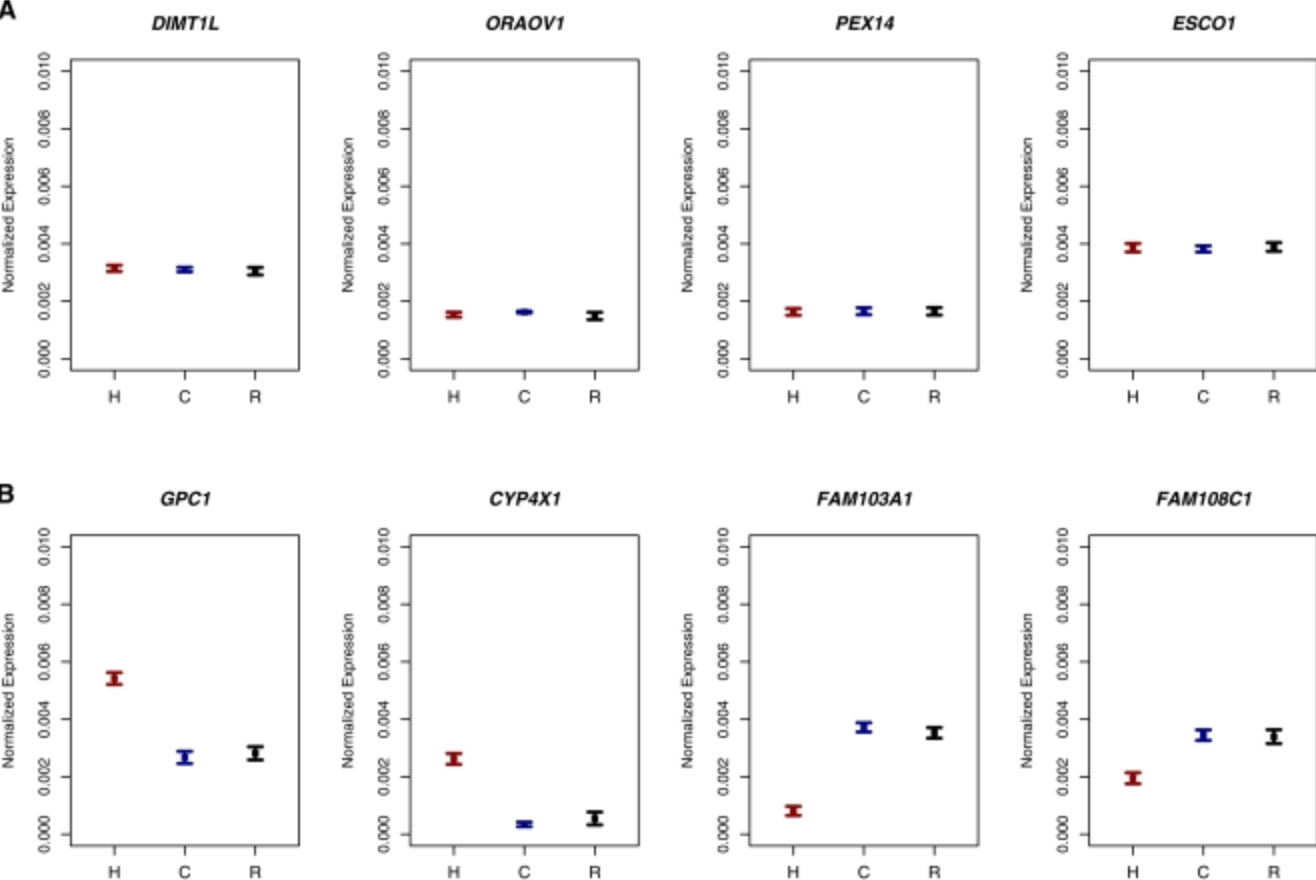
Sex-specific and lineage-specific alternative splicing in primates
Blekhman R, Marioni JC, Zumbo P, Stephens M, and Gilad Y
Genome Research; 2010
2009
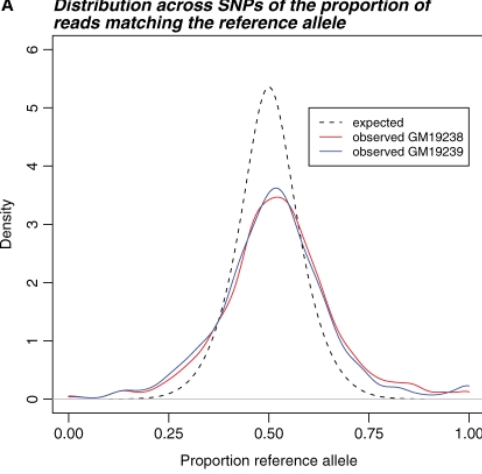
Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data
Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, and Pritchard JK
Bioinformatics; 2009
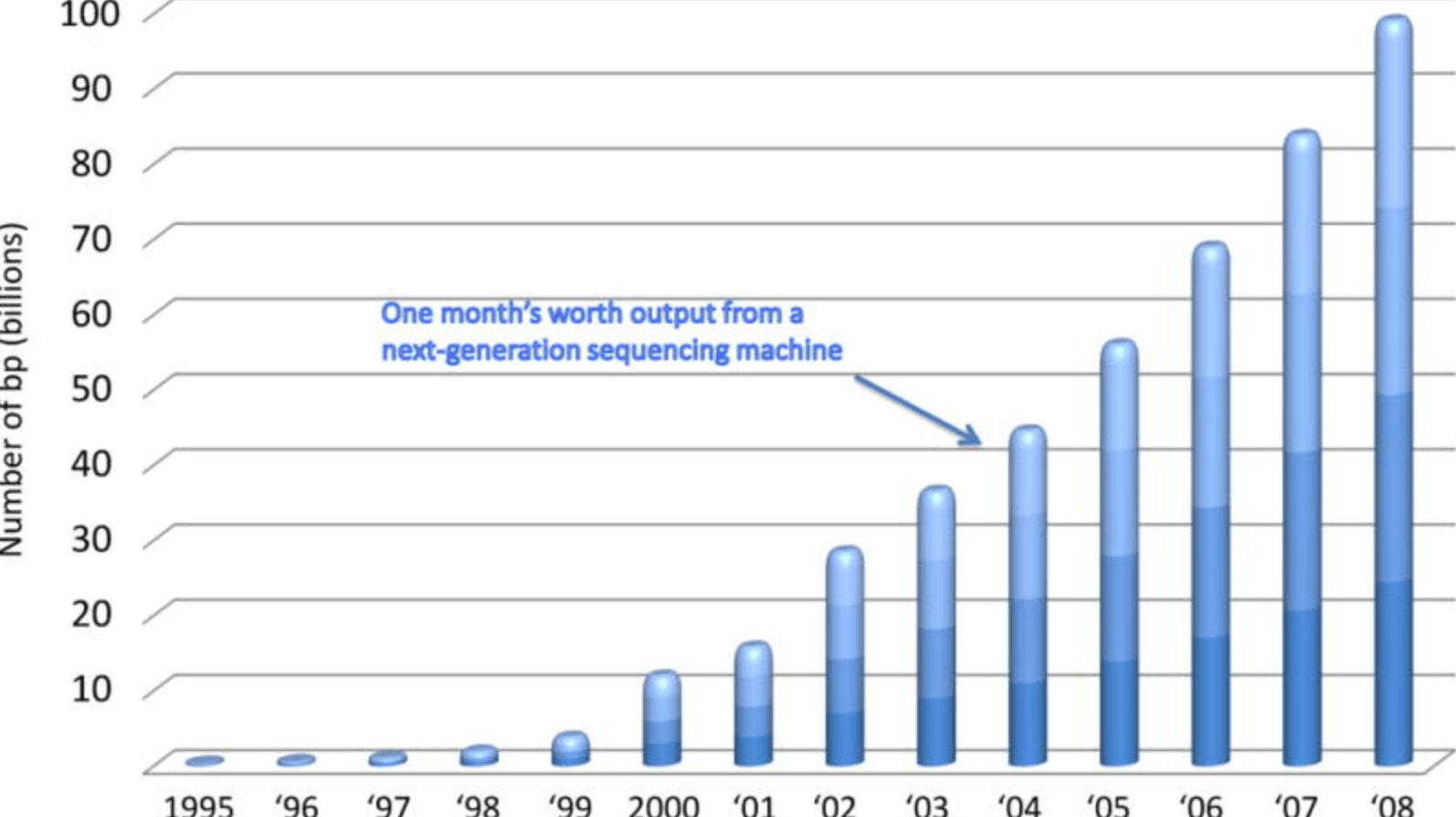
Characterizing natural variation using next-generation sequencing technologies
Gilad Y, Pritchard JK, and Thornton K
Trends in Genetics; 2009
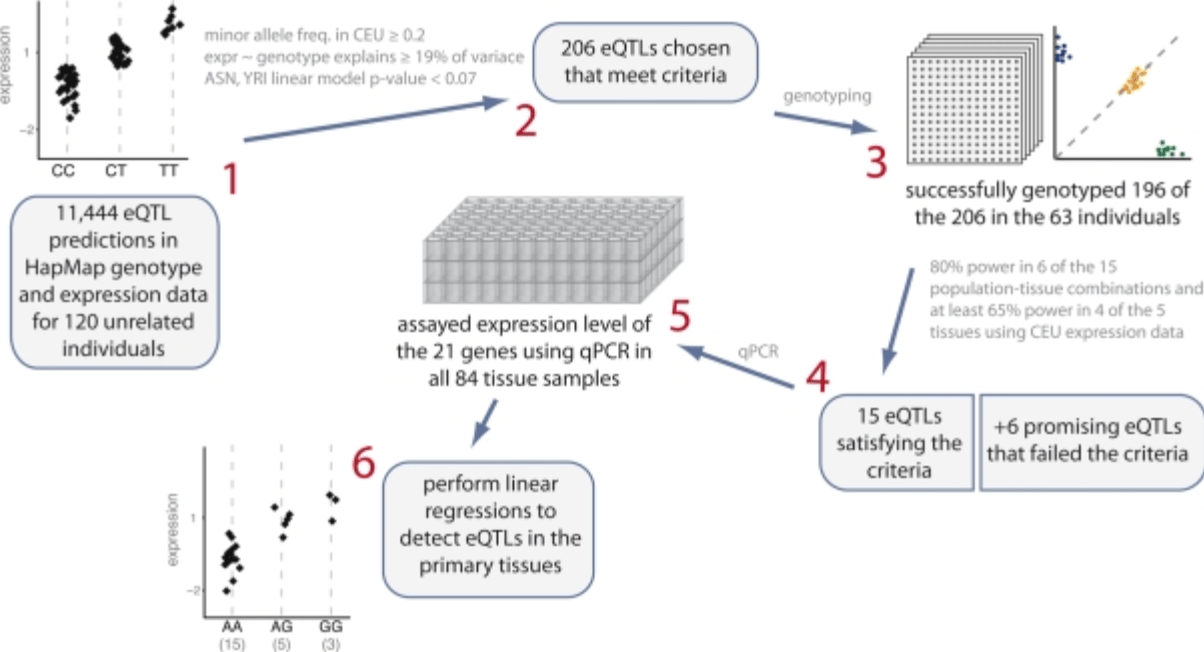
Expression quantitative trait loci detected in cell-lines are often present in primary tissues
Bullaughey K, Chavarria CI, Coop G, and Gilad Y
Human Molecular Genetics; 2009
Supplementary data: ZIP file


Copy number variation of CCL3-like genes affects rate of progression to simian-AIDS in rhesus macaques (Macaca mulatta)
Degenhardt JD, de Candia P, Chabot A, Schwartz S, Henderson L, Ling B, Hunter M, Jiang Z, Palermo RE, Katze M, Eichler EE, Ventura M, Rogers J, Marx P, Gilad Y, and Bustamante CD
PLoS Genetics; 2009

Olfactory receptor genes: Human loss during evolution. Squire LR (ed.)
Gilad Y
Encyclopedia of Neuroscience, Oxford: Academic Press 2009, volume 7: 157-161.
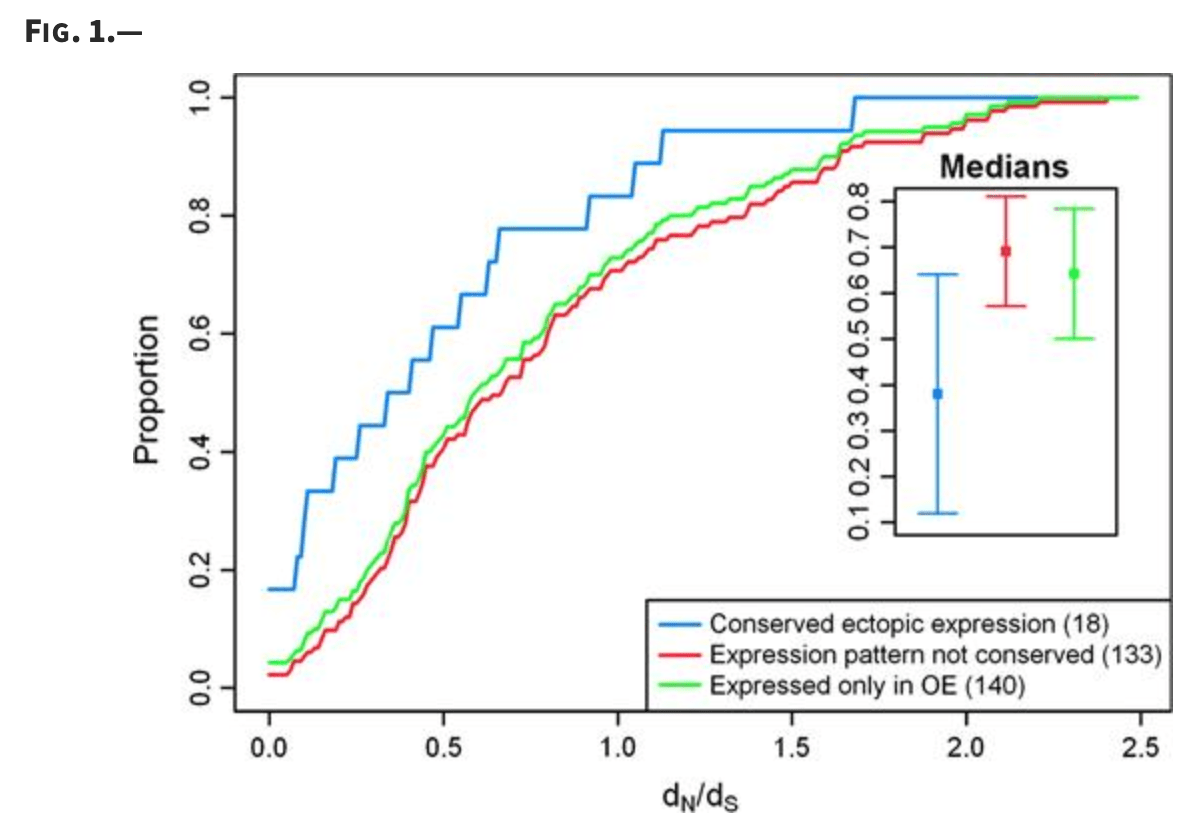
A signature of evolutionary constraint on a subset of ectopically expressed olfactory receptor genes
De la Cruz O, Blekhman R, Zhang X, Nicolae D, Firestein S, and Gilad Y
2008
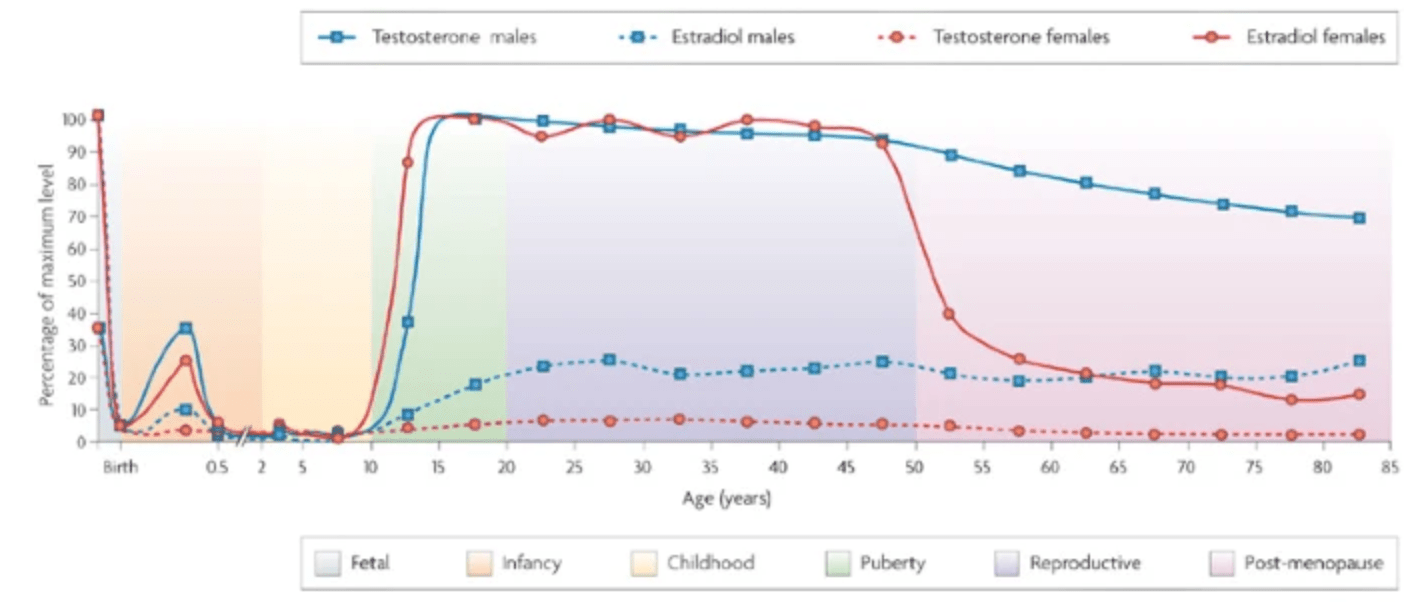
Sex-specific genetic architecture of human disease
Ober C, Loisel DA, and Gilad Y
Nature Reviews Genetics; 2008
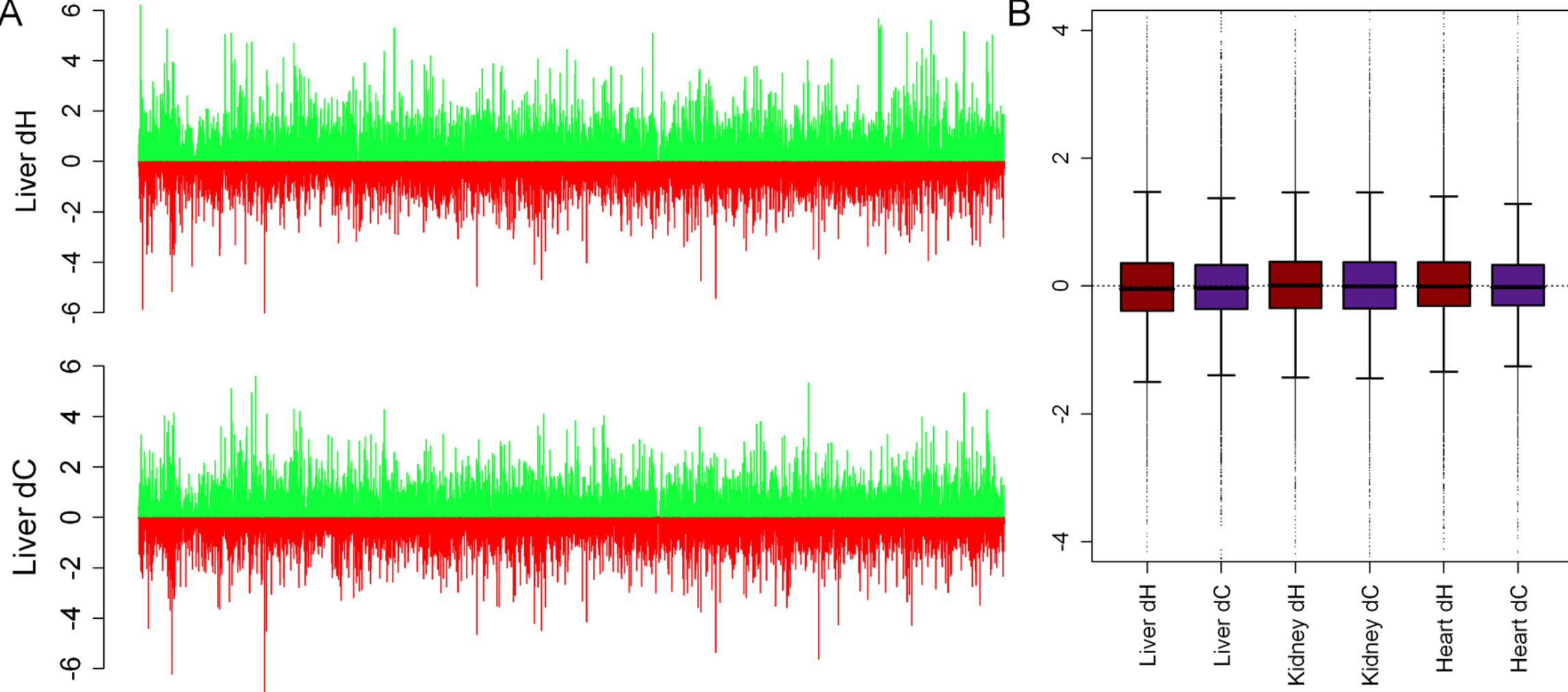
Gene regulation in primates evolves under tissue-specific selection pressures
Blekhman R, Oshlack A, Chabot AE, Smyth GK, and Gilad Y
PLoS Genetics; 2008
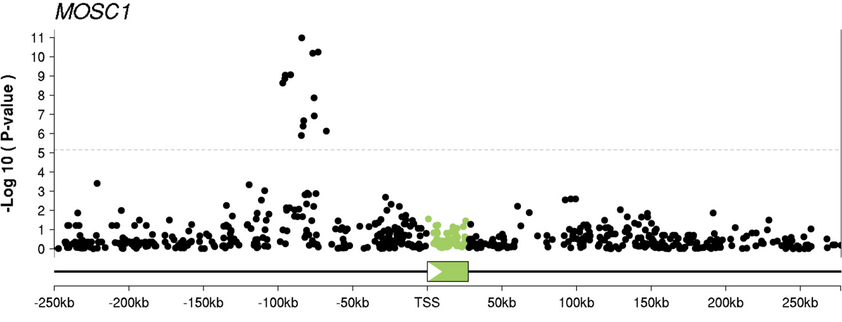
High-resolution mapping of expression-QTLs yields insight into human gene regulation
Veyrieras J-B, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, and Pritchard JK
PLoS Genetics; 2008
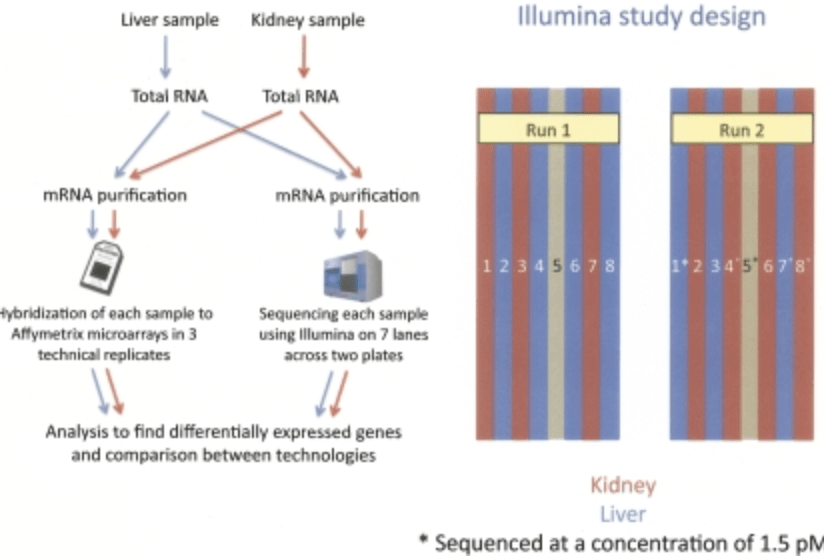
RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays
Marioni JC, Mason CE, Shrikant MM, Stephens M, and Gilad Y
Genome Research; 2008
Supplementary Data: Multiple file types
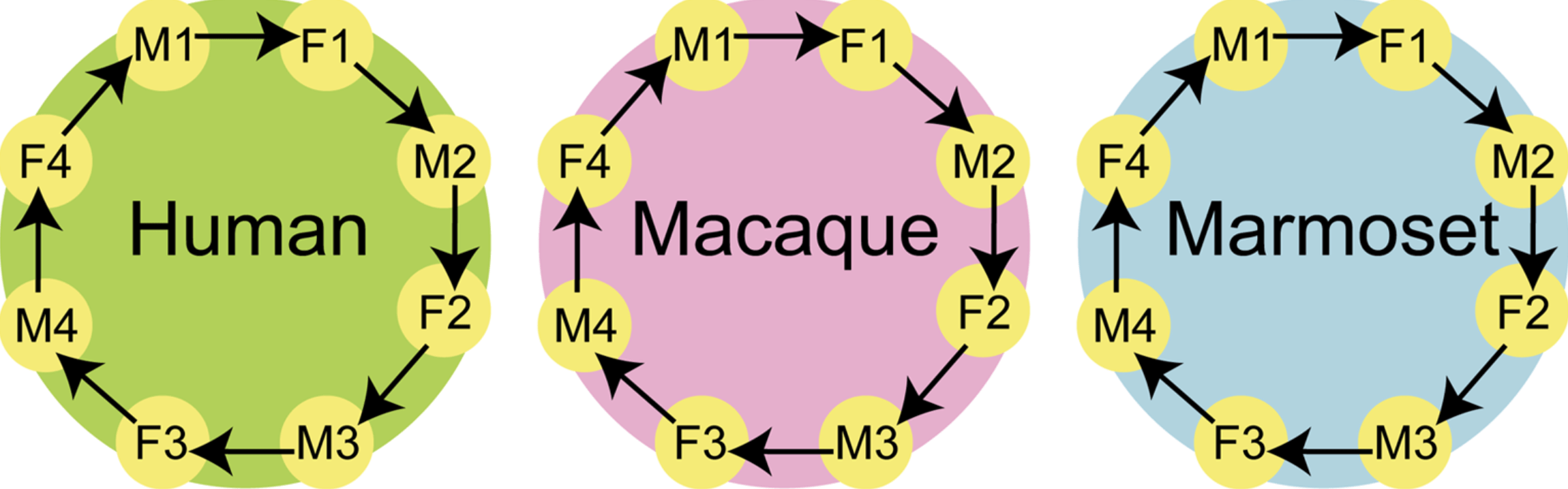
An evolutionarily conserved sexual signature in the primate brain
Reinius B, Saetre P, Leonard JA, Blekhman R, Martinez R, Gilad Y, and Jazin H
PLoS Genetics; 2008
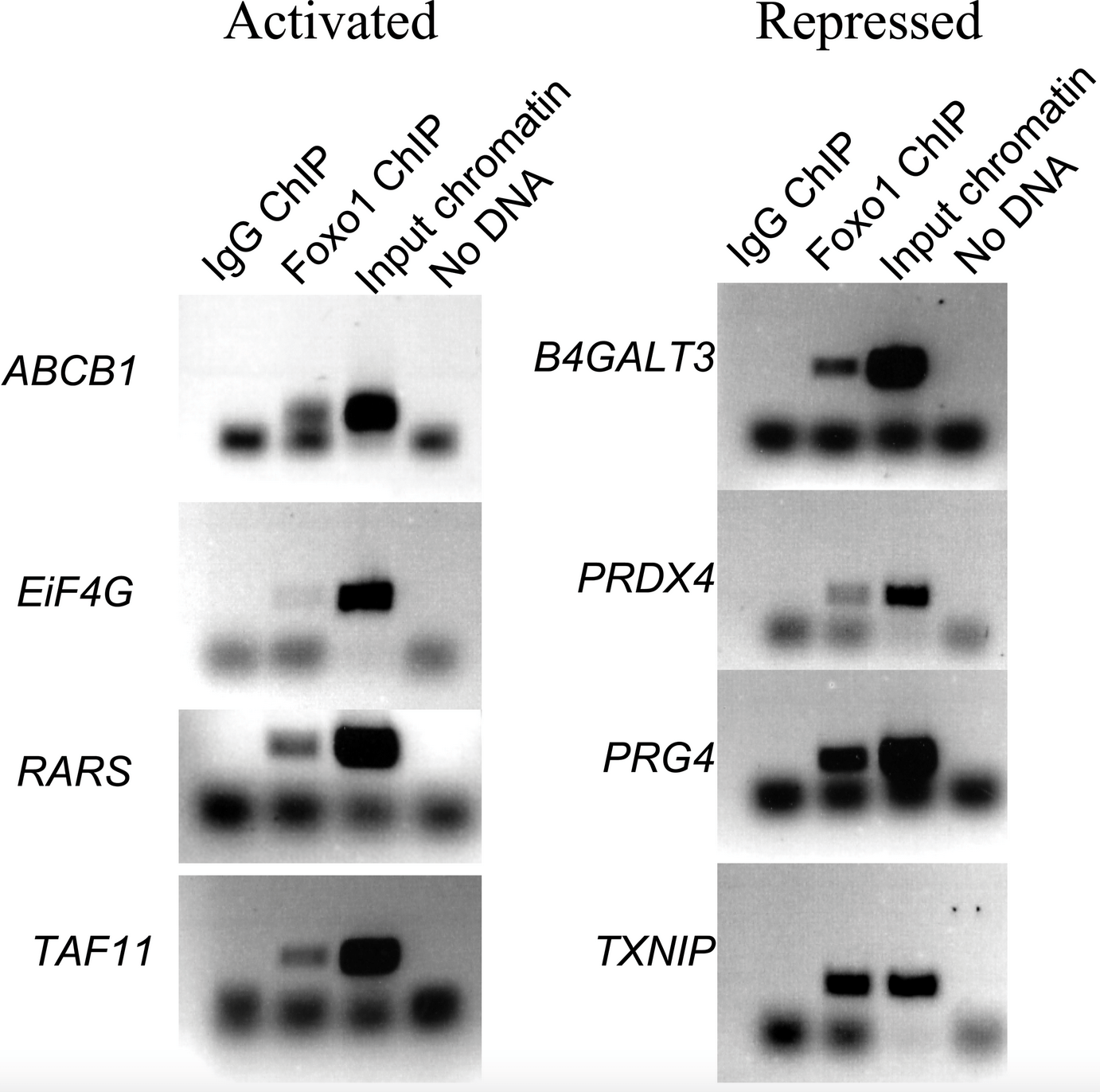
A Combination of Genomic Approaches Reveals the Role of FOXO1a in Regulating an Oxidative Stress Response Pathway
de Candia P, Blekhman R, Chabot AE, Oshlack A and Gilad Y
PLoS One; 2008
2007
A framework for exploring functional variability in olfactory receptor genes
Man O, Willhite DC, Crasto CJ, Shepherd GM and Gilad Y
PLoS One; 2007
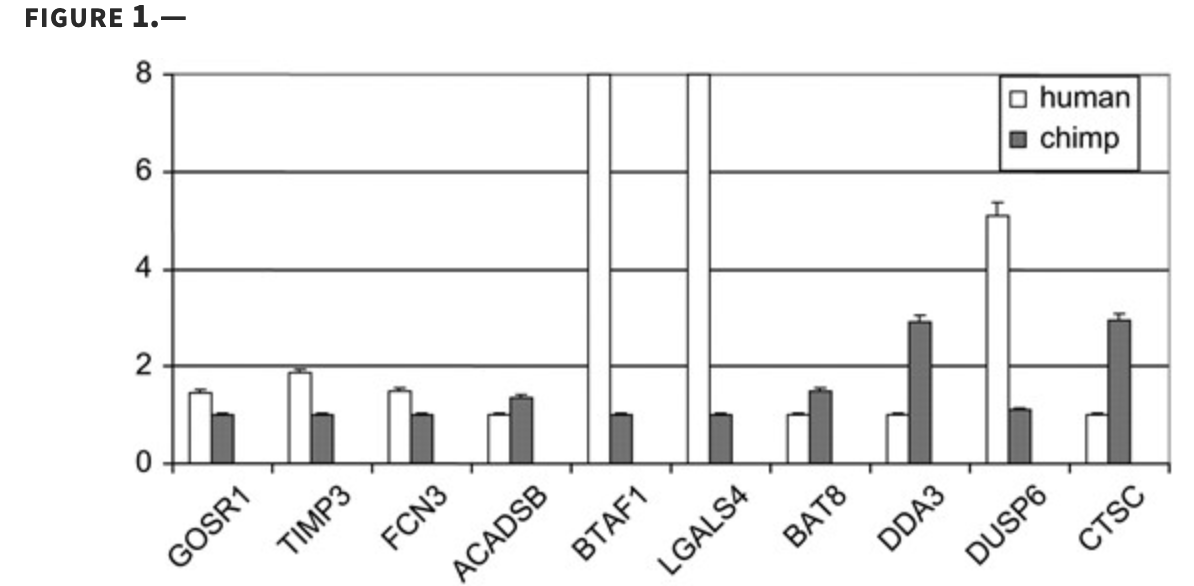
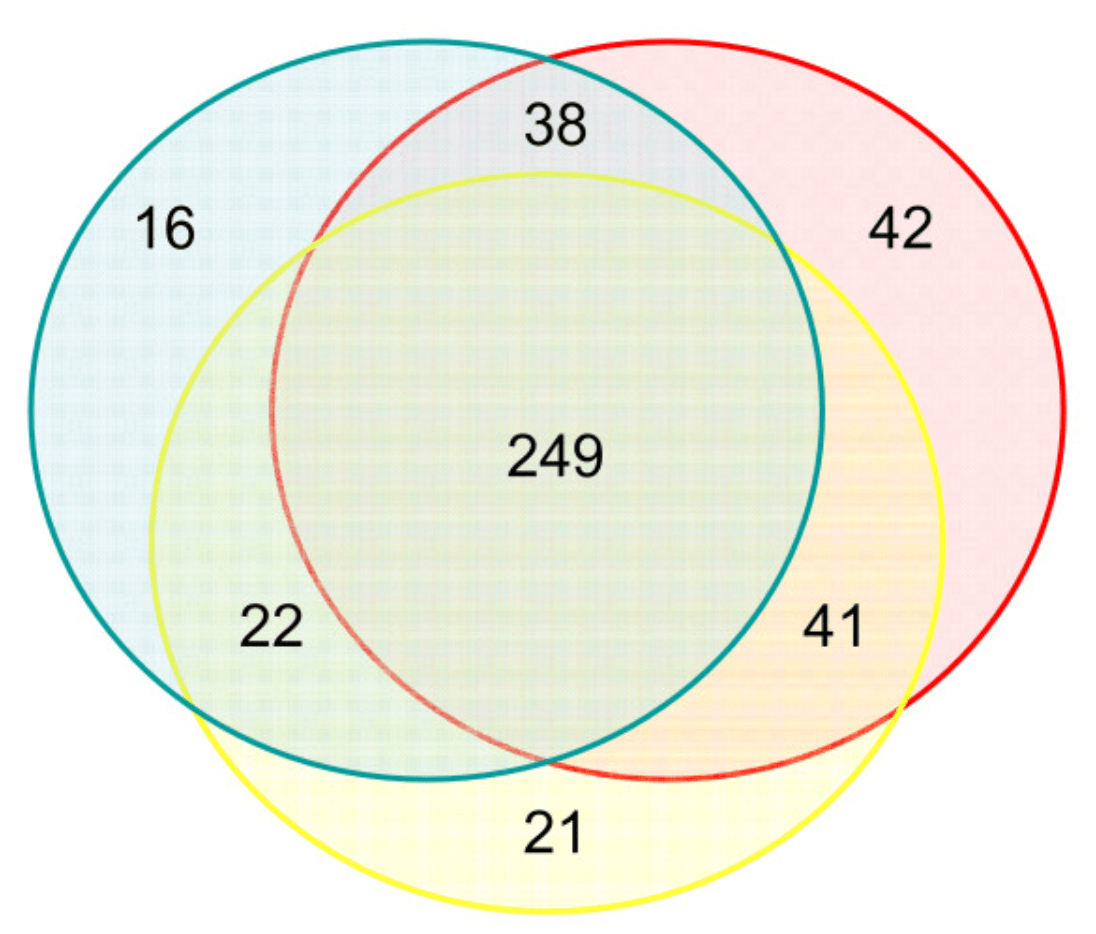
Characterizing the expression of human olfactory receptor gene family using a novel DNA microarray
Zhang X, De la Cruz O, Pinto JM, Nicolae D, Firestein S and Gilad Y
Genome Biology; 2007
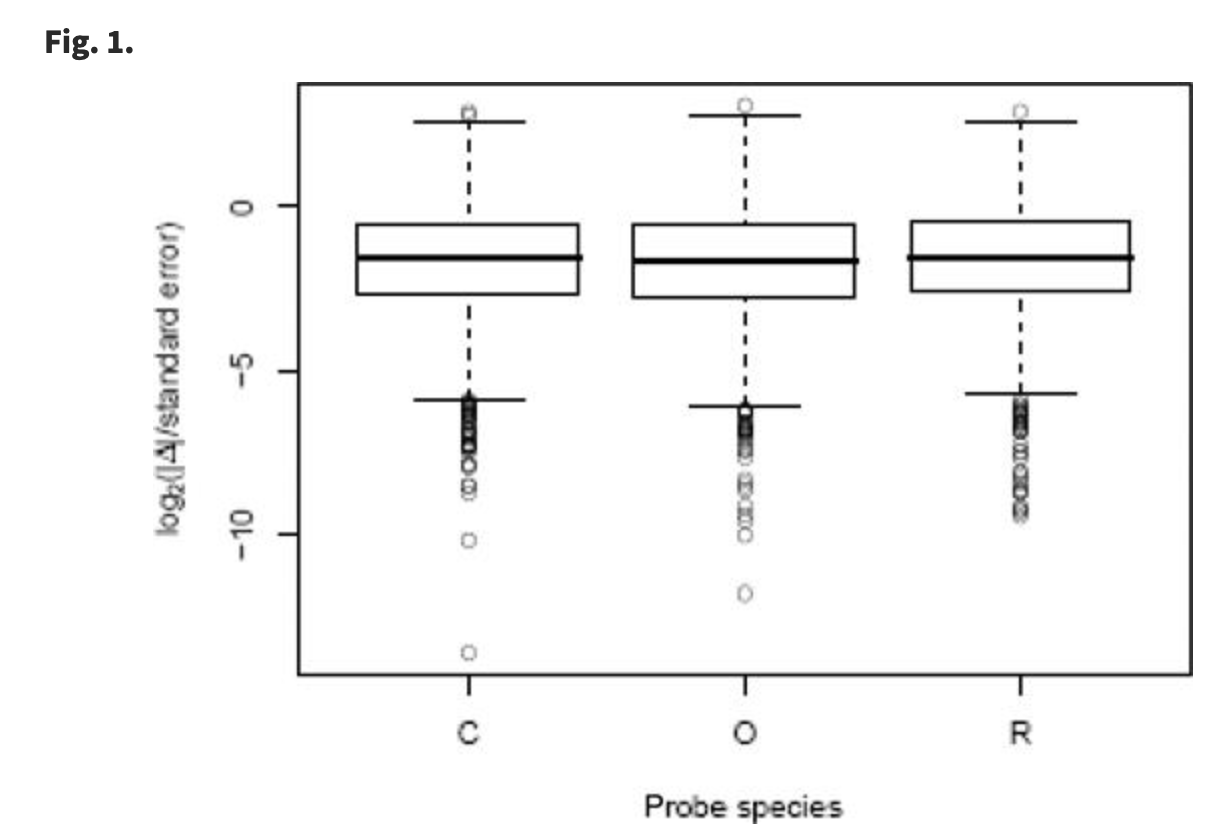
Using DNA microarrays to study gene expression in closely related species
Oshlack A, Chabot AE, Smyth GK and Gilad Y
Bioinformatics; 2007
Supplementary data: DOC file
2006
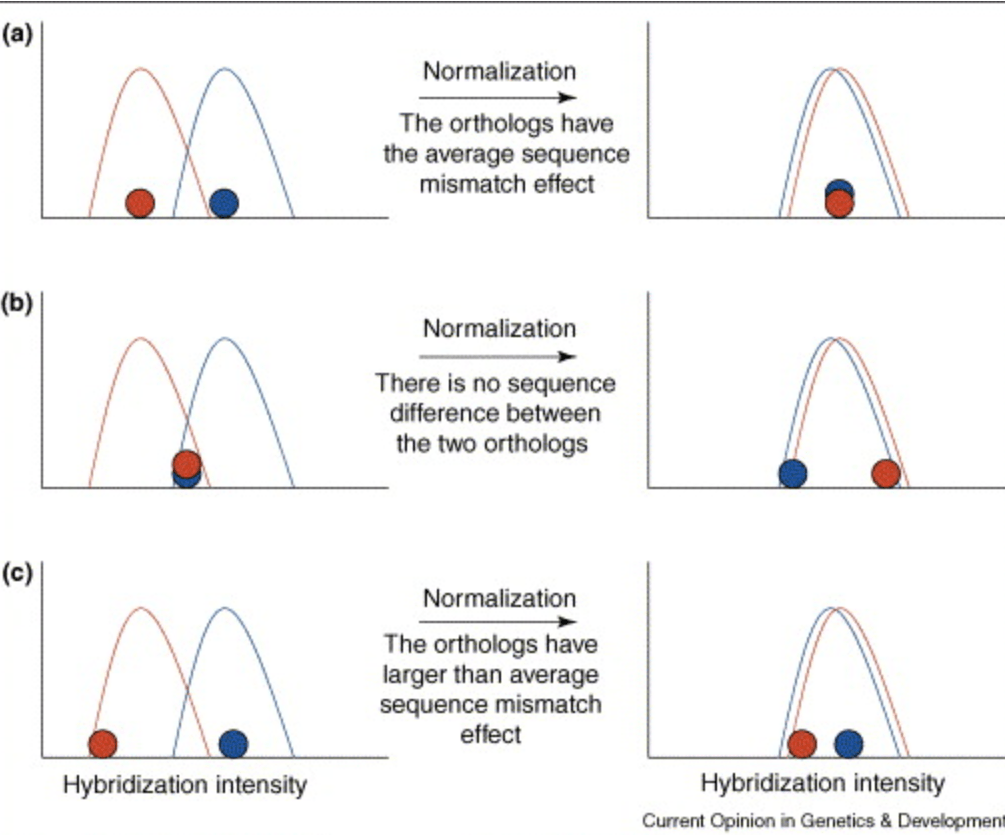
Using microarrays to study natural variation
Gilad Y and Borevitz J
Expression profiling in primates reveals a rapid evolution of human transcription factors
Gilad Y, Oshlack A, Smyth GK, Speed TP and White KP
Nature; 2006
2000-2005
Initial sequence of the chimpanzee genome and comparison with the human genome
The Chimpanzee Sequencing and Analysis Consortium
Nature; 2005
Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles
Gilad Y, Rifkin SA, Bertone P, Gerstein M and White KP
Genome Research; 2005
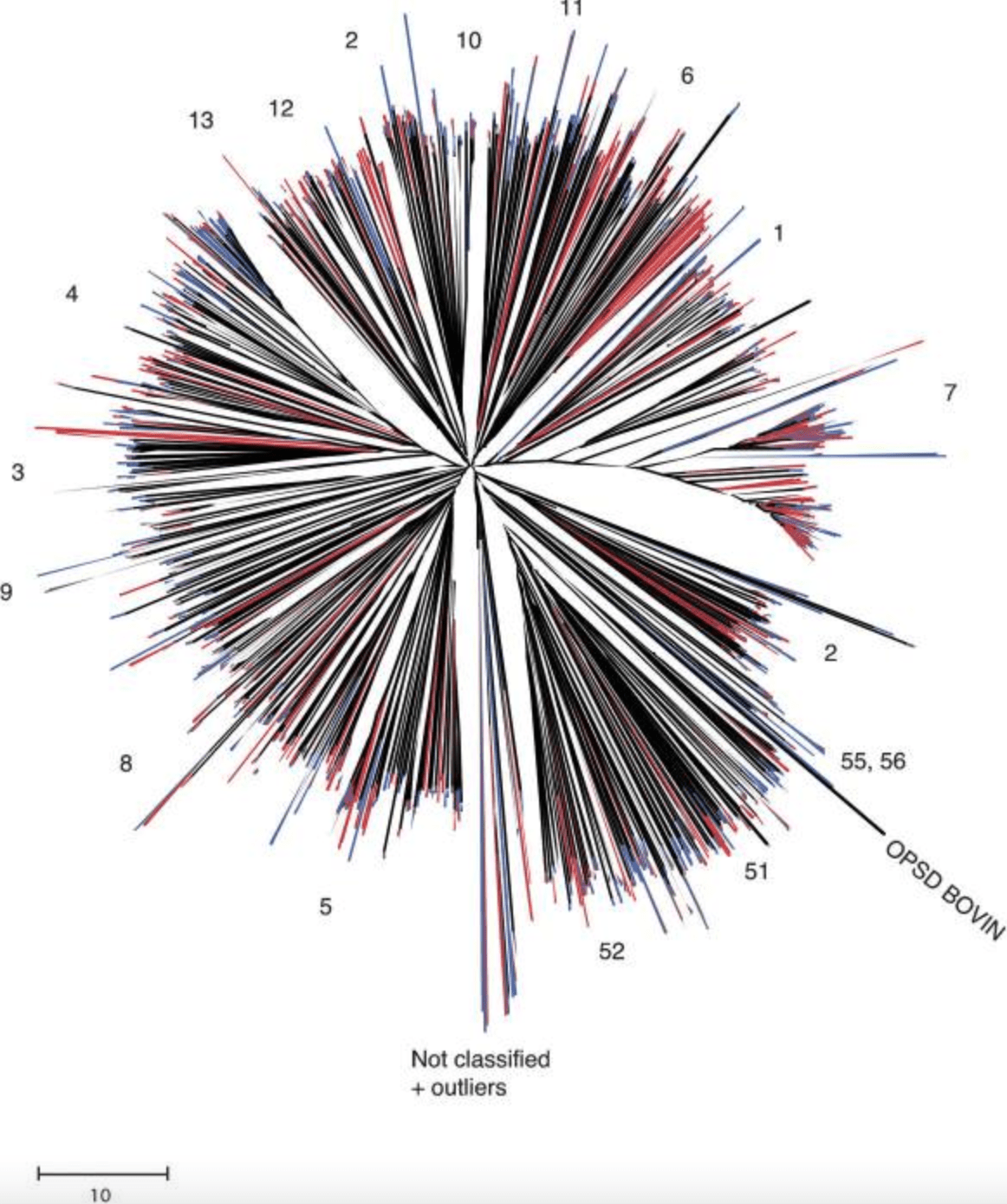
A comparison of the human and chimpanzee olfactory receptor gene repertoires
Gilad Y, Man O and Glusman G
Genome Research; 2005

Evolution of bitter taste receptors in humans and apes
Fischer A, Gilad Y, Man O and Pääbo S
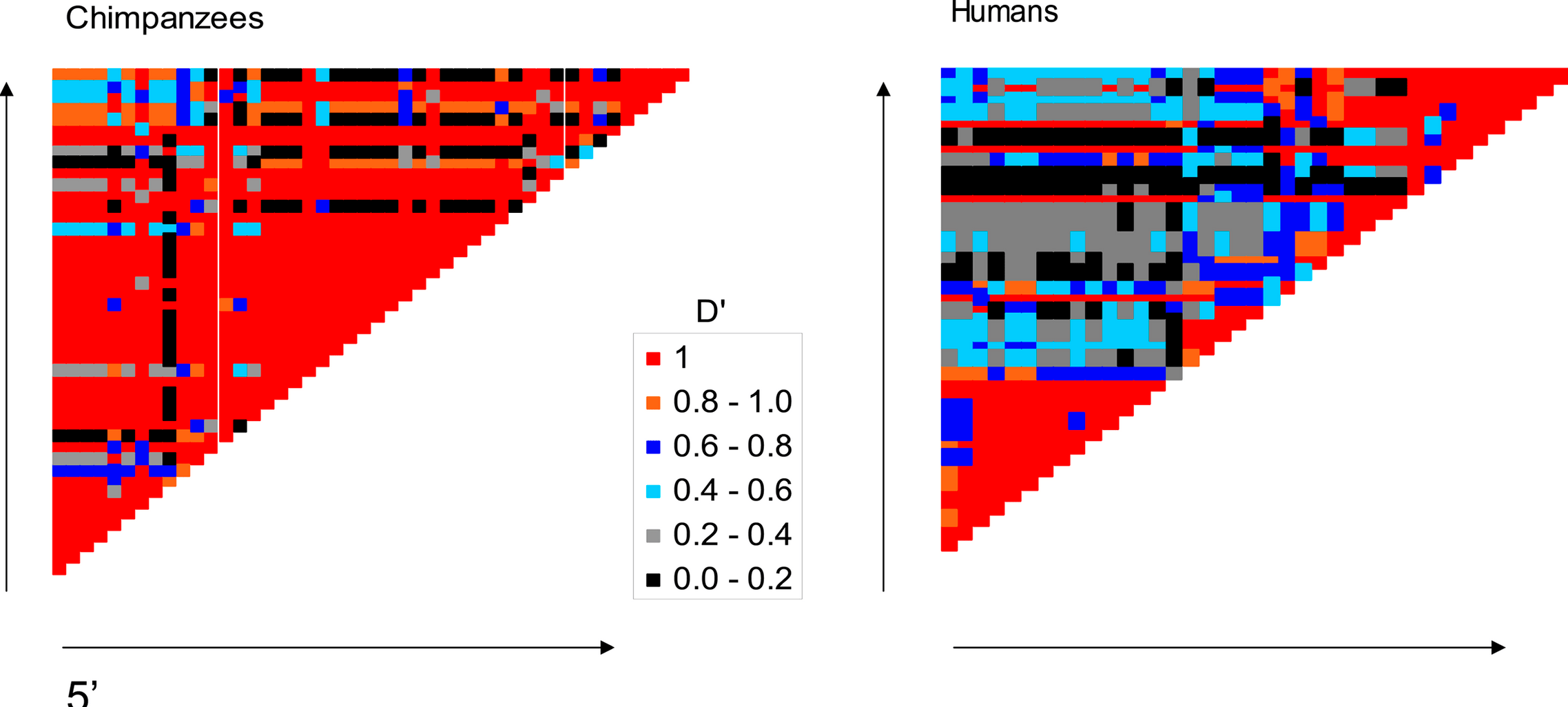
Absence of the TAP2 human recombination hotspot in chimpanzee
Ptak SE, Roeder AD, Stephens M, Gilad Y, Pääbo S and Przeworski, M
PLoS Biology; 2004

Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates
Gilad Y, Przeworski M, and Lancet D
PLoS Biology; 2004
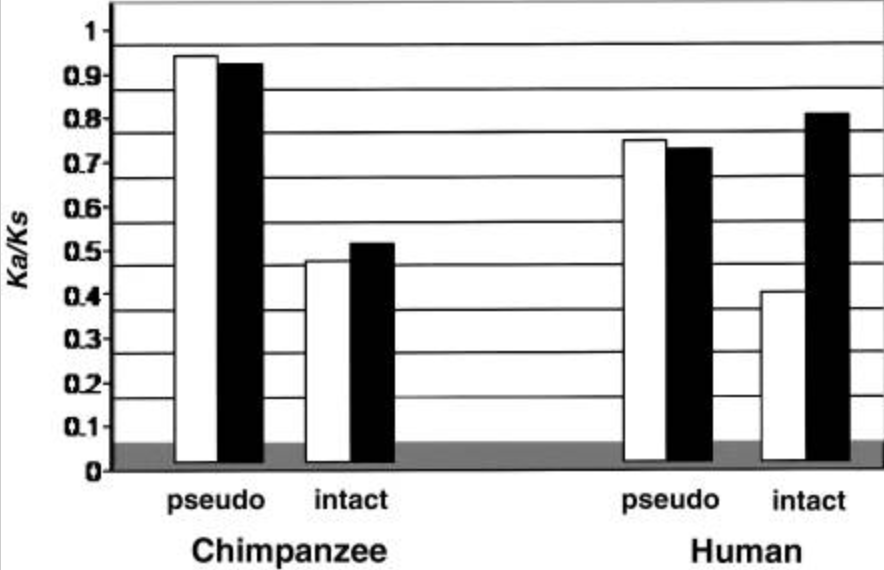
Natural selection on the olfactory receptor gene family in humans and chimpanzees
Gilad Y, Bustamante CD, Lancet D and Pääbo S
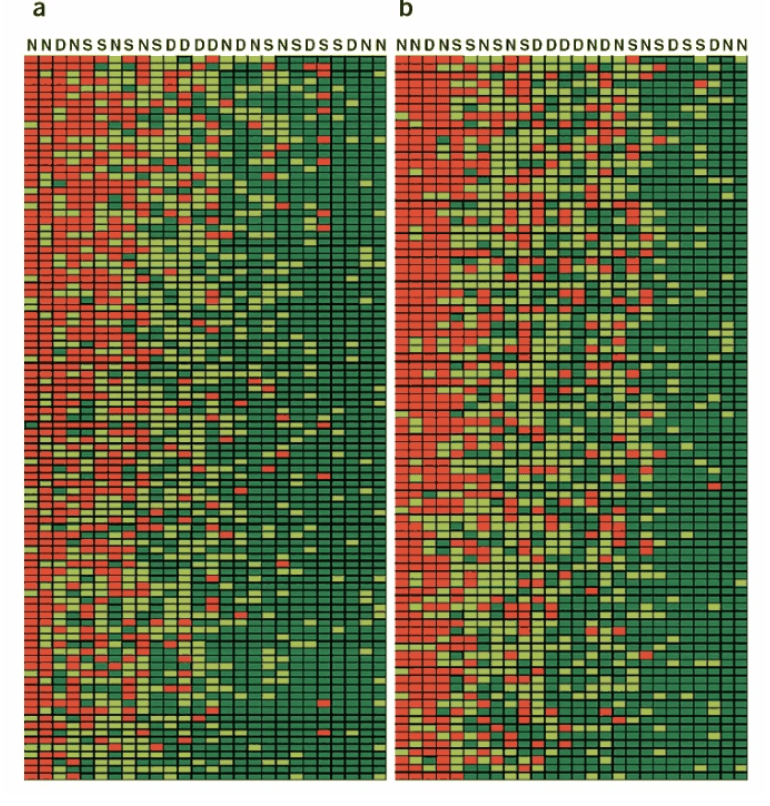
Population differences in the human functional olfactory repertoire
Gilad Y and Lancet D
Population differences in haplotype structure within a human olfactory receptor gene cluster
Menashe I, Man O, Lancet D and Gilad Y
Evidence for positive selection and population structure at the human MAO-A gene
Gilad Y, Rozenberg S, Przeworski M, Lancet D and Skorecki K
Mechanisms for evolving hypervariability: the case of conopeptides
Conticello GS, Gilad Y, Avidan N, Ben-Asher E, Levy Z and Fainzilber M
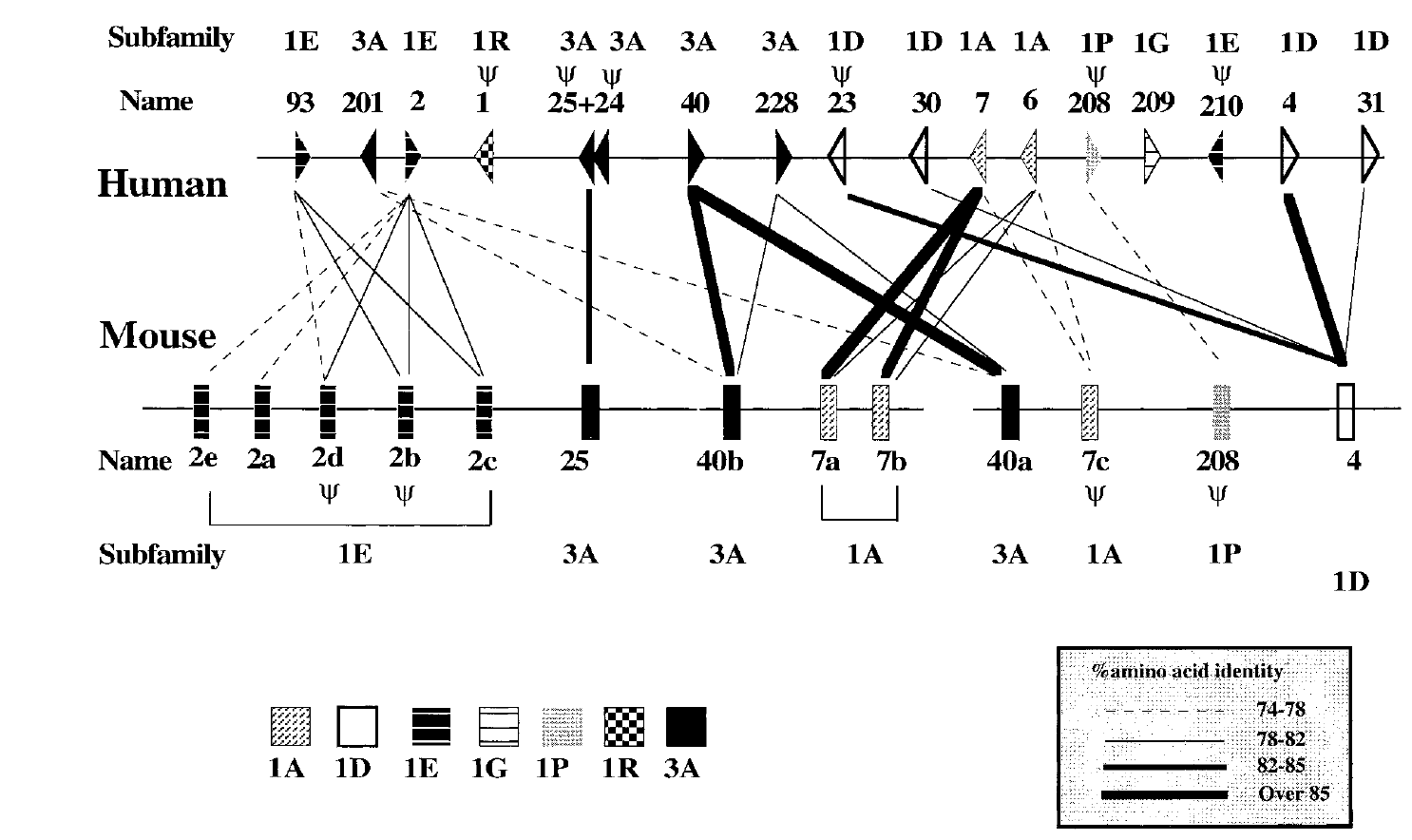
Mouse-Human Orthology relationships in an Olfactory Receptor Gene Cluster
Lapidot M, Pilpel T, Gilad Y, Falcovits A, Haaf T and Lancet D
Genomics; 2001
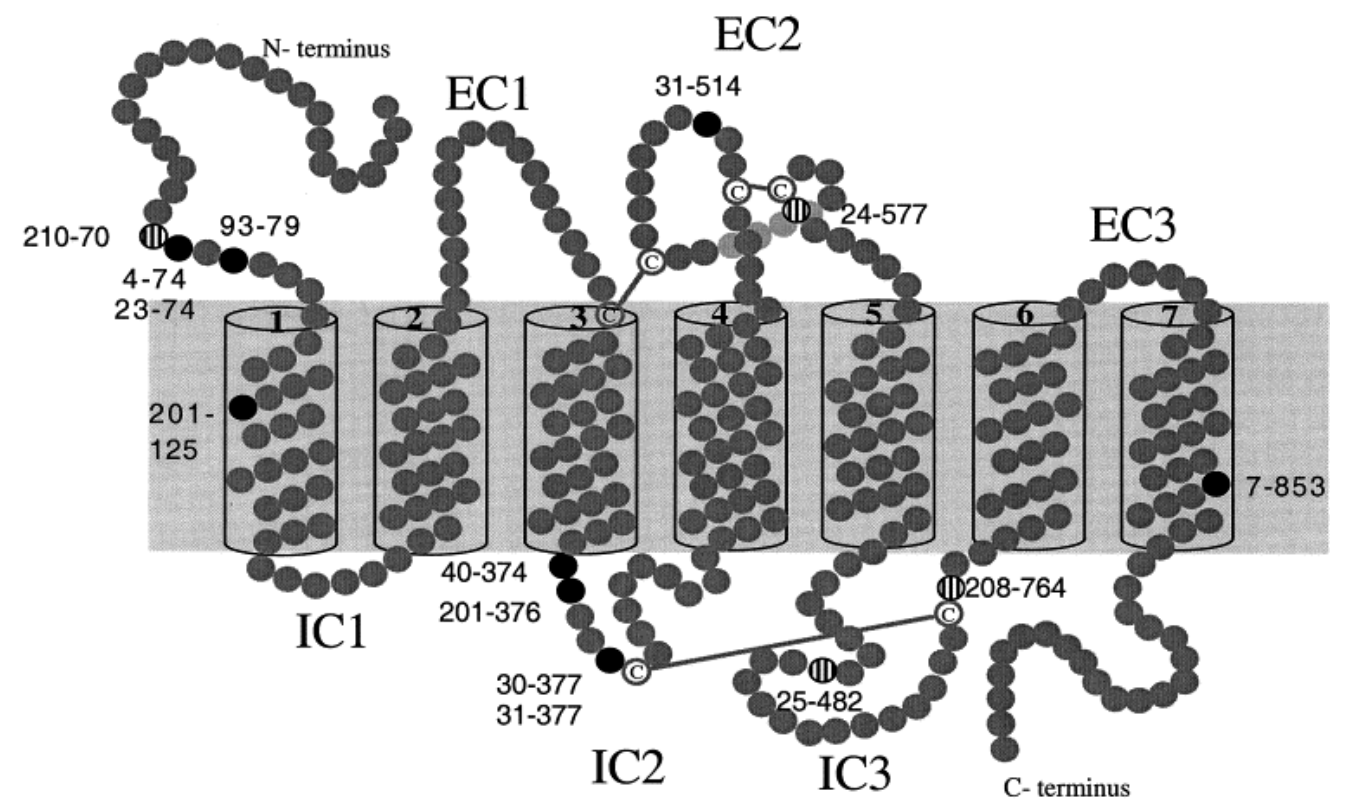
Dichotomy of single- nucleotide polymorphism haplotypes in olfactory receptor genes and pseudogenes
Gilad Y, Segré D, Skorecki K, Nachman MW, Lancet D and Sharon D
Nature Genetics; 2000
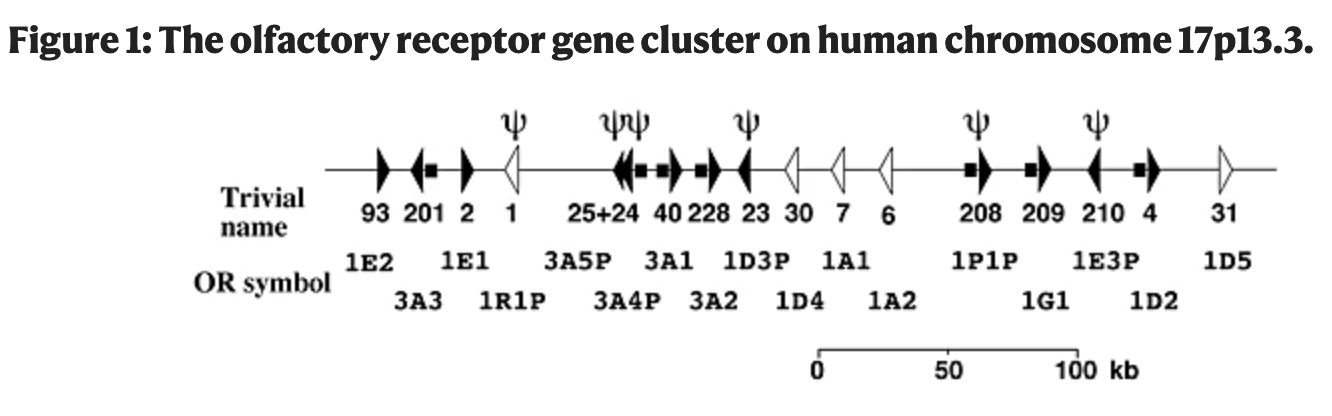
Identification and characterization of coding single-nucleotide polymorphisms within a human olfactory receptor gene cluster
Sharon D, Gilad Y, Glusman G, Khen M, Lancet D and Kalush F
Genes; 2000
